You are browsing environment: FUNGIDB
CAZyme Information: sr16207-t26_1-p1
You are here: Home > Sequence: sr16207-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporisorium reilianum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Ustilaginomycetes; ; Ustilaginaceae; Sporisorium; Sporisorium reilianum | |||||||||||
| CAZyme ID | sr16207-t26_1-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 76 | 295 | 2.6e-62 | 0.9553571428571429 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 371727 | Glyco_hydro_cc | 1.27e-74 | 78 | 298 | 16 | 234 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 431 | 1 | 431 | |
| 1.18e-309 | 1 | 431 | 1 | 431 | |
| 1.07e-255 | 1 | 423 | 1 | 421 | |
| 4.92e-212 | 47 | 405 | 40 | 404 | |
| 1.15e-206 | 42 | 408 | 42 | 421 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.76e-23 | 56 | 265 | 20 | 234 | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
|
| 9.69e-23 | 56 | 265 | 20 | 234 | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
|
| 2.50e-22 | 56 | 265 | 20 | 234 | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei] |
|
| 2.50e-22 | 56 | 265 | 20 | 234 | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose [Amycolatopsis mediterranei],6UAU_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose [Amycolatopsis mediterranei] |
|
| 1.79e-19 | 80 | 264 | 23 | 207 | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites [Blastomyces gilchristii SLH14081] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
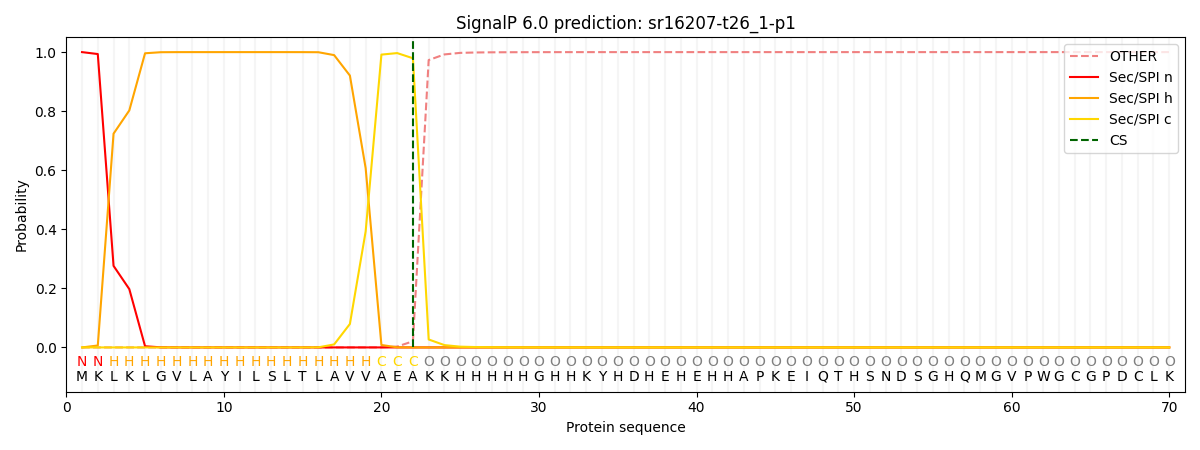
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000207 | 0.999752 | CS pos: 22-23. Pr: 0.9792 |
