You are browsing environment: FUNGIDB
CAZyme Information: sr15758-t26_1-p1
Basic Information
help
| Species |
Sporisorium reilianum
|
| Lineage |
Basidiomycota; Ustilaginomycetes; ; Ustilaginaceae; Sporisorium; Sporisorium reilianum
|
| CAZyme ID |
sr15758-t26_1-p1
|
| CAZy Family |
GT2 |
| CAZyme Description |
related to cellulase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 470 |
FQ311438|CGC1 |
52447.00 |
5.8712 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_SreilianumSRZ2 |
6791 |
999809 |
118 |
6673
|
|
| Gene Location |
No EC number prediction in sr15758-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
43 |
420 |
1.4e-133 |
0.9970326409495549 |
sr15758-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
470 |
1 |
470 |
| 0.0 |
1 |
470 |
1 |
470 |
| 6.05e-274 |
1 |
470 |
1 |
468 |
| 7.60e-215 |
1 |
470 |
1 |
367 |
| 2.99e-145 |
61 |
439 |
2 |
376 |
sr15758-t26_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.18e-33 |
35 |
452 |
30 |
388 |
Glycosyl hydrolase 5 family protein OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
| 3.99e-09 |
24 |
266 |
26 |
239 |
Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
This protein is predicted as SP
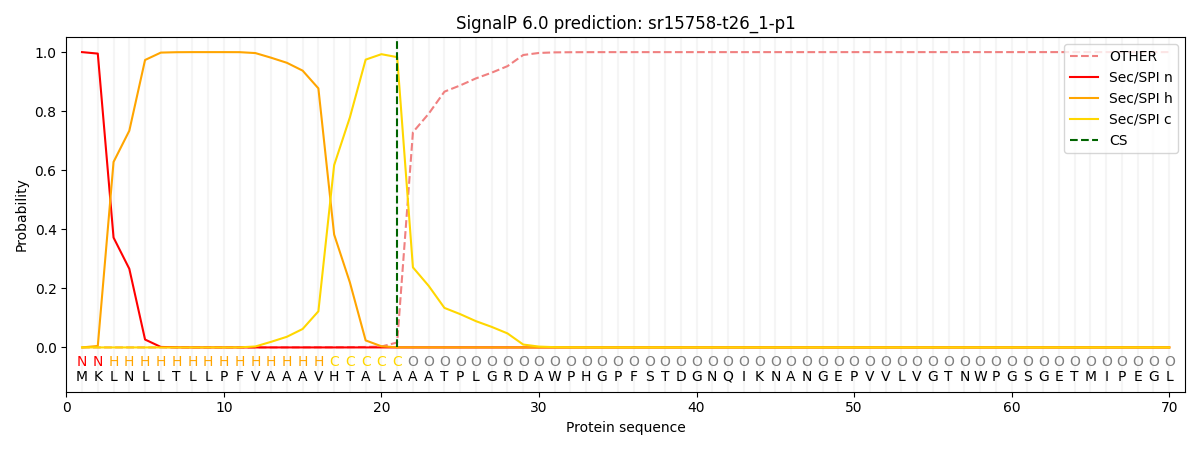
| Other |
SP_Sec_SPI |
CS Position |
| 0.000210 |
0.999739 |
CS pos: 21-22. Pr: 0.9830 |
There is no transmembrane helices in sr15758-t26_1-p1.

