You are browsing environment: FUNGIDB
CAZyme Information: sr14846-t26_1-p1
You are here: Home > Sequence: sr14846-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporisorium reilianum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Ustilaginomycetes; ; Ustilaginaceae; Sporisorium; Sporisorium reilianum | |||||||||||
| CAZyme ID | sr14846-t26_1-p1 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 902 | 1352 | 4.4e-16 | 0.35964912280701755 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397353 | Glyco_hydro_63 | 2.34e-08 | 980 | 1355 | 185 | 488 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| 282904 | Herpes_BLLF1 | 3.01e-07 | 80 | 363 | 460 | 796 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
| 411345 | gliding_GltJ | 2.09e-05 | 103 | 261 | 392 | 521 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 225805 | DamX | 3.98e-05 | 110 | 282 | 39 | 228 | Cell division protein DamX, binds to the septal ring, contains C-terminal SPOR domain [Cell cycle control, cell division, chromosome partitioning]. |
| 235906 | PRK07003 | 5.62e-05 | 103 | 360 | 402 | 629 | DNA polymerase III subunit gamma/tau. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1413 | 1 | 1413 | |
| 0.0 | 1 | 1413 | 1 | 1418 | |
| 0.0 | 1 | 1413 | 1 | 1392 | |
| 0.0 | 1 | 1413 | 1 | 1419 | |
| 0.0 | 1 | 1413 | 1 | 1379 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 362 | 1374 | 12 | 953 | Uncharacterized protein YMR196W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YMR196W PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
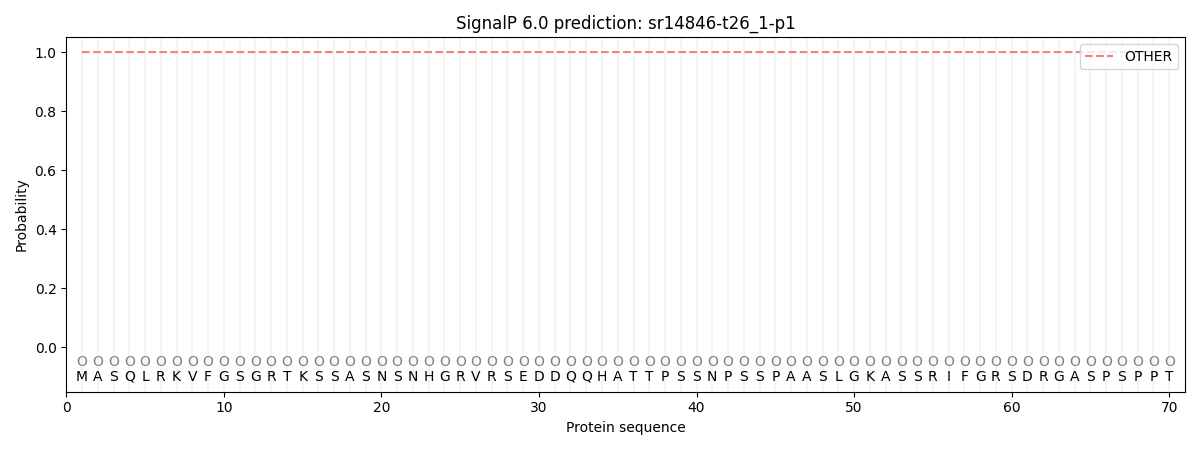
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000058 | 0.000000 |
