You are browsing environment: FUNGIDB
CAZyme Information: sr13229-t26_1-p1
You are here: Home > Sequence: sr13229-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Sporisorium reilianum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Ustilaginomycetes; ; Ustilaginaceae; Sporisorium; Sporisorium reilianum | |||||||||||
| CAZyme ID | sr13229-t26_1-p1 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | probable Endoglucanase 1 precursor (egl1) | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH45 | 27 | 235 | 2e-62 | 0.9949494949494949 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 280233 | Glyco_hydro_45 | 2.30e-101 | 26 | 235 | 1 | 214 | Glycosyl hydrolase family 45. |
| 411231 | MSCRAMM_ClfA | 6.27e-14 | 278 | 478 | 688 | 888 | MSCRAMM family adhesin clumping factor ClfA. Clumping factor A is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). It is heavily studied in Staphylococcus aureus both for its biological role in adhesion and for its potential for vaccination. Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| 411231 | MSCRAMM_ClfA | 6.01e-11 | 295 | 459 | 563 | 727 | MSCRAMM family adhesin clumping factor ClfA. Clumping factor A is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). It is heavily studied in Staphylococcus aureus both for its biological role in adhesion and for its potential for vaccination. Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| 411414 | MSCRAMM_ClfB | 2.43e-08 | 278 | 449 | 642 | 813 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| 411414 | MSCRAMM_ClfB | 5.43e-08 | 278 | 446 | 654 | 824 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.37e-211 | 1 | 282 | 1 | 282 | |
| 1.86e-208 | 1 | 282 | 1 | 283 | |
| 3.19e-193 | 1 | 280 | 1 | 280 | |
| 5.37e-176 | 1 | 282 | 1 | 279 | |
| 5.35e-164 | 1 | 282 | 1 | 288 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.78e-53 | 24 | 236 | 19 | 221 | Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus [Cryptopygus antarcticus],5H4U_B Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus [Cryptopygus antarcticus],5H4U_C Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus [Cryptopygus antarcticus] |
|
| 9.17e-45 | 24 | 246 | 23 | 233 | Apo Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A],6MVJ_A Cellobiose complex Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A] |
|
| 1.03e-44 | 24 | 246 | 2 | 212 | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126],5GM9_A Crystal structure of a glycoside hydrolase in complex with cellobiose [Thermothielavioides terrestris NRRL 8126] |
|
| 1.16e-44 | 24 | 246 | 5 | 215 | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126] |
|
| 8.20e-43 | 27 | 245 | 3 | 209 | Structure of Melanocarpus albomyces endoglucanase in complex with cellobiose [Melanocarpus albomyces],1OA9_A Structure of Melanocarpus albomyces endoglucanase [Melanocarpus albomyces] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.53e-177 | 1 | 282 | 1 | 279 | Endoglucanase 1 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=EGL1 PE=2 SV=1 |
|
| 2.48e-52 | 24 | 236 | 19 | 221 | Endoglucanase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
|
| 5.76e-46 | 3 | 234 | 14 | 240 | Endoglucanase OS=Phaedon cochleariae OX=80249 PE=2 SV=1 |
|
| 9.38e-44 | 9 | 252 | 6 | 237 | Putative endoglucanase type K OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 8.24e-41 | 27 | 244 | 3 | 209 | Endoglucanase-5 OS=Humicola insolens OX=34413 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
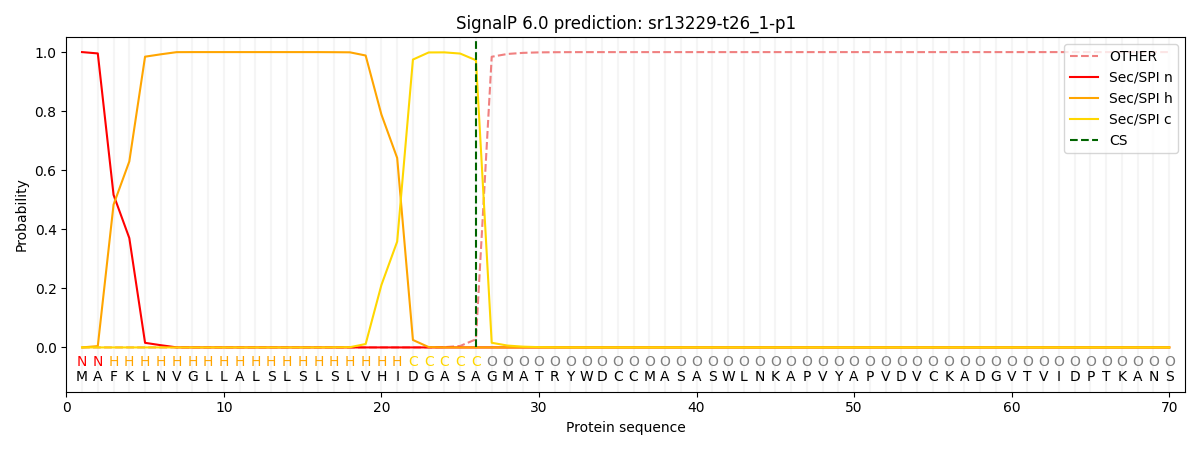
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000203 | 0.999797 | CS pos: 26-27. Pr: 0.9725 |
