You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_008874-t41_1-p1
You are here: Home > Sequence: jhhlp_008874-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
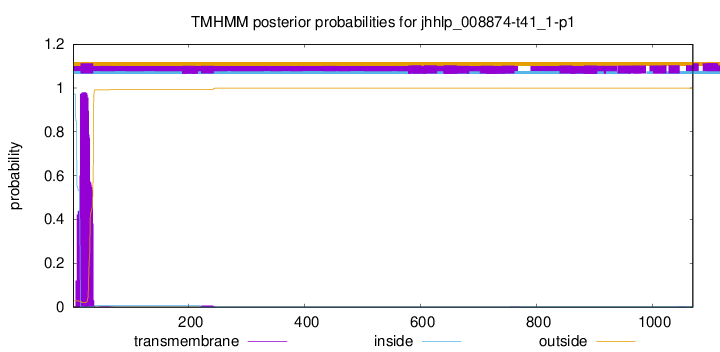
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_008874-t41_1-p1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE18 | 258 | 627 | 6.1e-164 | 0.989100817438692 |
| CBM87 | 36 | 255 | 6.5e-98 | 0.9908256880733946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397000 | ComA | 2.96e-104 | 808 | 1053 | 1 | 237 | (2R)-phospho-3-sulfolactate synthase (ComA). In methanobacteria (2R)-phospho-3-sulfolactate synthase (ComA) catalyzes the first step of the biosynthesis of coenzyme M from phosphoenolpyruvate (P-enolpyruvate). This novel enzyme catalyzes the stereospecific Michael addition of sulfite to P-enolpyruvate, forming L-2-phospho-3-sulfolactate (PSL). It is suggested that the ComA-catalyzed reaction is analogous to those reactions catalyzed by beta-elimination enzymes that proceed through an enolate intermediate. |
| 224722 | ComA | 7.52e-65 | 809 | 1066 | 10 | 256 | Phosphosulfolactate synthase, CoM biosynthesis protein A [Coenzyme transport and metabolism]. |
| 213023 | CE4_NodB_like_5s_6s | 0.005 | 398 | 457 | 71 | 123 | Putative catalytic NodB homology domain of PgaB, IcaB, and similar proteins which consist of a deformed (beta/alpha)8 barrel fold with 5- or 6-strands. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes bacterial poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB, hemin storage system HmsF protein in gram-negative species, intercellular adhesion proteins IcaB, and many uncharacterized prokaryotic polysaccharide deacetylases. It also includes a putative polysaccharide deacetylase YxkH encoded by the Bacillus subtilis yxkH gene, which is one of six polysaccharide deacetylase gene homologs present in the Bacillus subtilis genome. Sequence comparison shows all family members contain a conserved domain similar to the catalytic NodB homology domain of rhizobial NodB-like proteins, which consists of a deformed (beta/alpha)8 barrel fold with 6 or 7 strands. However, in this family, most proteins have 5 strands and some have 6 strands. Moreover, long insertions are found in many family members, whose function remains unknown. |
| 200589 | CE4_GLA_like_6s | 0.009 | 398 | 493 | 60 | 152 | Putative catalytic NodB homology domain of gellan lyase and similar proteins. This family is represented by the extracellular polysaccharide-degrading enzyme, gellan lyase (gellanase, EC 4.2.2.-), from Bacillus sp. The enzyme acts on gellan exolytically and releases a tetrasaccharide of glucuronyl-glucosyl-rhamnosyl-glucose with unsaturated glucuronic acid at the nonreducing terminus. The family also includes many uncharacterized prokaryotic polysaccharide deacetylases, which show high sequence similarity to Bacillus sp. gellan lyase. Although their biological functions remain unknown, all members of the family contain a conserved domain with a 6-stranded beta/alpha barrel, which is similar to the catalytic NodB homology domain of rhizobial NodB-like proteins, belonging to the larger carbohydrate esterase 4 (CE4) superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.59e-275 | 30 | 700 | 366 | 1029 | |
| 1.06e-268 | 27 | 700 | 42 | 708 | |
| 3.21e-265 | 23 | 700 | 25 | 699 | |
| 5.50e-264 | 14 | 700 | 14 | 691 | |
| 1.04e-261 | 13 | 700 | 11 | 691 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.38e-231 | 27 | 700 | 22 | 684 | Crystal structure of Agd3 a novel carbohydrate deacetylase [Aspergillus fumigatus Af293] |
|
| 5.38e-15 | 833 | 1066 | 26 | 251 | Crystal structure of Methanococcus jannaschii phosphosulfolactate synthase [Methanocaldococcus jannaschii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.17e-62 | 804 | 1053 | 10 | 267 | Protein HEAT-STRESS-ASSOCIATED 32 OS=Arabidopsis thaliana OX=3702 GN=HSA32 PE=2 SV=1 |
|
| 1.77e-19 | 800 | 1066 | 2 | 256 | Phosphosulfolactate synthase OS=Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H) OX=187420 GN=comA PE=3 SV=1 |
|
| 2.77e-14 | 833 | 1066 | 26 | 251 | Phosphosulfolactate synthase OS=Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) OX=243232 GN=comA PE=1 SV=1 |
|
| 2.97e-11 | 813 | 1056 | 27 | 261 | Phosphosulfolactate synthase OS=Xanthobacter autotrophicus (strain ATCC BAA-1158 / Py2) OX=78245 GN=xecG PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
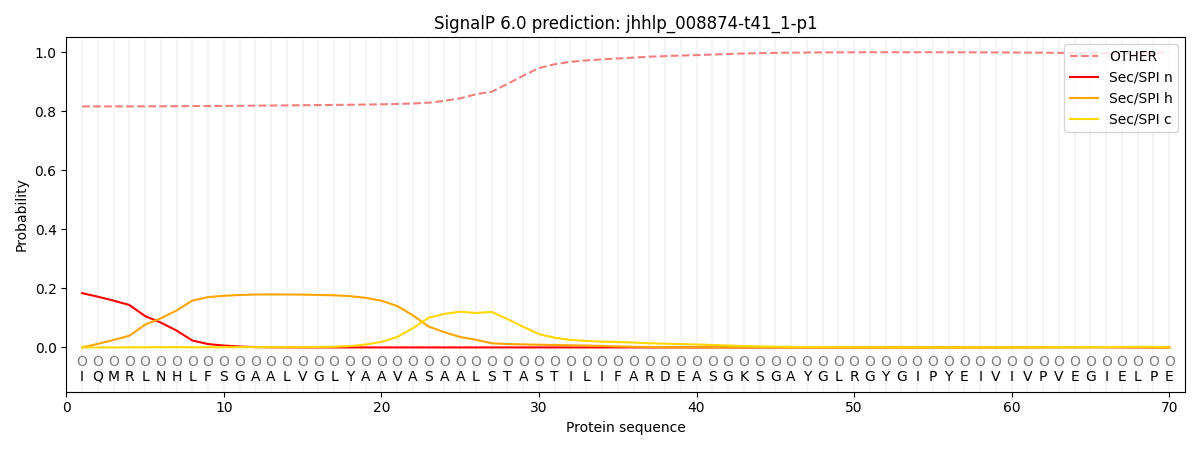
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.825945 | 0.174071 |