You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_006063-t41_1-p1
You are here: Home > Sequence: jhhlp_006063-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_006063-t41_1-p1 | |||||||||||
| CAZy Family | GH43|CBM91 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.99.18:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 146 | 694 | 1.9e-235 | 0.9872262773722628 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 3.38e-64 | 159 | 693 | 9 | 535 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 2.08e-36 | 159 | 692 | 7 | 532 | choline dehydrogenase; Validated |
| 398739 | GMC_oxred_C | 2.90e-27 | 553 | 687 | 3 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 366272 | GMC_oxred_N | 3.07e-18 | 238 | 459 | 23 | 217 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 215420 | PLN02785 | 1.02e-12 | 153 | 692 | 51 | 577 | Protein HOTHEAD |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.09e-254 | 148 | 694 | 239 | 785 | |
| 3.66e-253 | 158 | 694 | 249 | 786 | |
| 6.43e-253 | 158 | 695 | 277 | 814 | |
| 2.02e-252 | 155 | 694 | 246 | 785 | |
| 2.02e-248 | 149 | 694 | 235 | 783 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.96e-230 | 158 | 694 | 8 | 545 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 2.75e-229 | 158 | 693 | 230 | 765 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
|
| 3.68e-227 | 158 | 694 | 230 | 767 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 1.64e-130 | 158 | 694 | 3 | 537 | Chain A, Cellobiose dehydrogenase [Phanerodontia chrysosporium],1NAA_B Chain B, Cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 1.91e-130 | 158 | 694 | 8 | 542 | Chain A, cellobiose dehydrogenase [Phanerodontia chrysosporium],1KDG_B Chain B, cellobiose dehydrogenase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.65e-127 | 158 | 694 | 235 | 769 | Cellobiose dehydrogenase OS=Phanerodontia chrysosporium OX=2822231 GN=CDH-1 PE=1 SV=1 |
|
| 1.65e-27 | 158 | 692 | 5 | 531 | Oxygen-dependent choline dehydrogenase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=betA PE=3 SV=1 |
|
| 5.24e-27 | 158 | 692 | 5 | 531 | Oxygen-dependent choline dehydrogenase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=betA PE=3 SV=1 |
|
| 8.96e-27 | 150 | 702 | 38 | 585 | Choline dehydrogenase, mitochondrial OS=Rattus norvegicus OX=10116 GN=Chdh PE=1 SV=1 |
|
| 4.25e-26 | 158 | 691 | 5 | 527 | L-sorbose 1-dehydrogenase OS=Gluconobacter oxydans OX=442 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
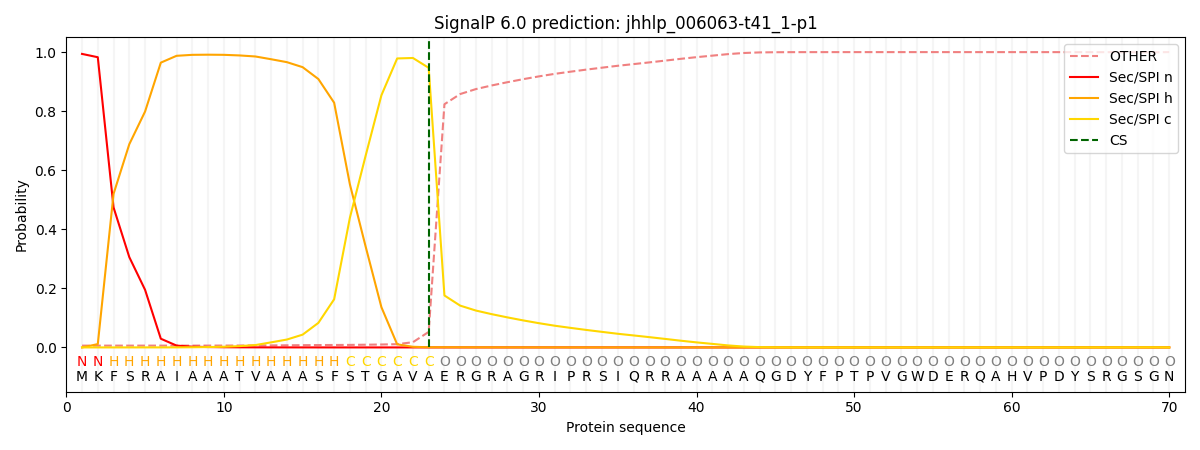
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.009473 | 0.990483 | CS pos: 23-24. Pr: 0.9468 |
