You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_005925-t41_1-p1
You are here: Home > Sequence: jhhlp_005925-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_005925-t41_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.-:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 105 | 428 | 2e-92 | 0.9888475836431226 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395595 | CBM_1 | 1.98e-15 | 19 | 46 | 2 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 5.32e-14 | 19 | 50 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 224423 | DAP2 | 0.003 | 229 | 346 | 435 | 550 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| 398876 | AXE1 | 0.009 | 253 | 295 | 161 | 203 | Acetyl xylan esterase (AXE1). This family consists of several bacterial acetyl xylan esterase proteins. Acetyl xylan esterases are enzymes that hydrolyze the ester linkages of the acetyl groups in position 2 and/or 3 of the xylose moieties of natural acetylated xylan from hardwood. These enzymes are one of the accessory enzymes which are part of the xylanolytic system, together with xylanases, beta-xylosidases, alpha-arabinofuranosidases and methylglucuronidases; these are all required for the complete hydrolysis of xylan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.00e-214 | 6 | 459 | 13 | 480 | |
| 4.00e-214 | 6 | 459 | 13 | 480 | |
| 5.36e-212 | 6 | 459 | 13 | 480 | |
| 6.41e-173 | 4 | 460 | 8 | 496 | |
| 3.26e-163 | 6 | 459 | 11 | 465 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.60e-144 | 90 | 459 | 10 | 374 | Chain A, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
|
| 1.30e-141 | 19 | 459 | 8 | 457 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 1.04e-136 | 102 | 459 | 56 | 410 | Crystal structure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
|
| 2.95e-136 | 102 | 459 | 56 | 410 | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile determined at 1.9 A resolution [Thermothelomyces thermophilus ATCC 42464],4G4J_A Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution [Thermothelomyces thermophilus ATCC 42464] |
|
| 7.83e-129 | 102 | 459 | 15 | 379 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.11e-215 | 6 | 459 | 13 | 480 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
|
| 1.14e-173 | 4 | 460 | 8 | 496 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Sodiomyces alcalophilus OX=398408 PE=1 SV=1 |
|
| 8.23e-160 | 4 | 459 | 7 | 459 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
|
| 1.43e-143 | 2 | 459 | 3 | 473 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
|
| 7.65e-143 | 1 | 459 | 1 | 471 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.18.61.1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
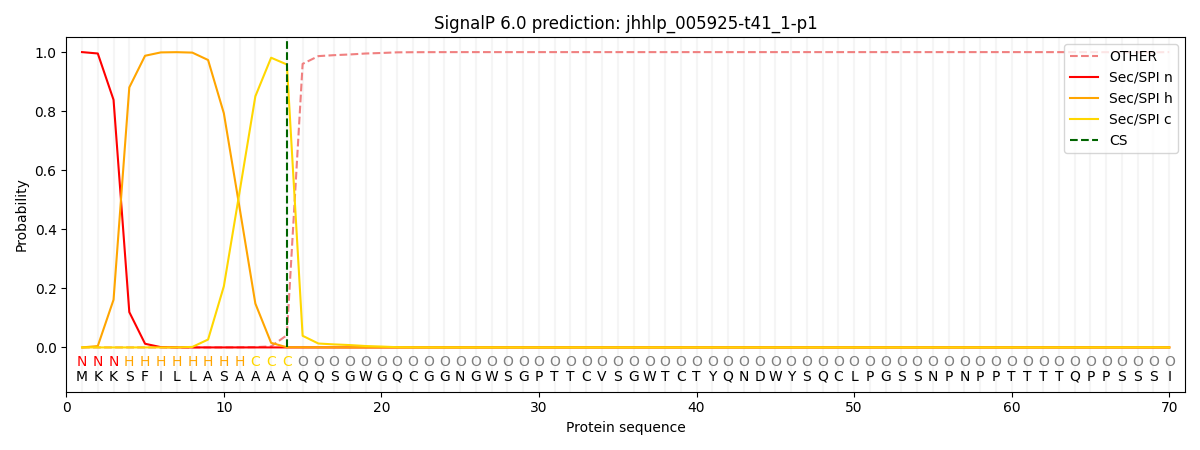
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002075 | 0.997900 | CS pos: 14-15. Pr: 0.9581 |
