You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_005277-t41_1-p1
You are here: Home > Sequence: jhhlp_005277-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_005277-t41_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.72:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 39 | 197 | 3.7e-21 | 0.6431718061674009 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 1.09e-25 | 8 | 168 | 18 | 180 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 402228 | Esterase_phd | 1.24e-22 | 42 | 237 | 5 | 194 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| 226584 | COG4099 | 1.12e-06 | 124 | 165 | 261 | 302 | Predicted peptidase [General function prediction only]. |
| 224423 | DAP2 | 4.70e-05 | 38 | 166 | 380 | 507 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| 223730 | Aes | 0.004 | 22 | 152 | 48 | 172 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.68e-138 | 9 | 295 | 9 | 293 | |
| 1.10e-131 | 13 | 360 | 9 | 363 | |
| 1.10e-131 | 13 | 360 | 9 | 363 | |
| 1.10e-131 | 13 | 360 | 9 | 363 | |
| 2.57e-127 | 22 | 295 | 32 | 306 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.60e-120 | 24 | 293 | 3 | 273 | Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.49e-128 | 4 | 349 | 13 | 353 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
|
| 4.56e-128 | 22 | 295 | 32 | 306 | Probable acetylxylan esterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=axeA PE=3 SV=1 |
|
| 2.62e-127 | 22 | 295 | 32 | 306 | Acetylxylan esterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=axeA PE=1 SV=1 |
|
| 2.62e-127 | 22 | 295 | 32 | 306 | Probable acetylxylan esterase A OS=Aspergillus flavus OX=5059 GN=axeA PE=3 SV=1 |
|
| 1.70e-125 | 4 | 348 | 13 | 349 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
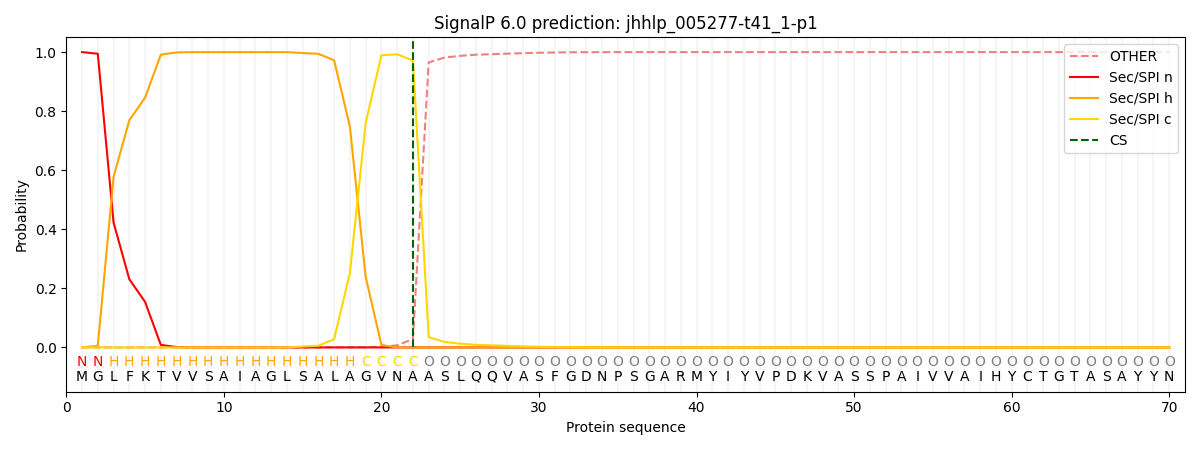
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000247 | 0.999753 | CS pos: 22-23. Pr: 0.9714 |
