You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_005005-t41_1-p1
You are here: Home > Sequence: jhhlp_005005-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_005005-t41_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.71:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH162 | 33 | 528 | 7e-203 | 0.9958677685950413 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 6 | 530 | 9 | 528 | |
| 3.94e-306 | 1 | 529 | 1 | 522 | |
| 1.60e-305 | 1 | 529 | 1 | 522 | |
| 6.50e-305 | 1 | 529 | 1 | 522 | |
| 9.23e-305 | 1 | 529 | 1 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.34e-236 | 32 | 529 | 13 | 505 | The apo-structure of endo-beta-1,2-glucanase from Talaromyces funiculosus [Talaromyces funiculosus],6IMU_B The apo-structure of endo-beta-1,2-glucanase from Talaromyces funiculosus [Talaromyces funiculosus],6IMV_A The complex structure of endo-beta-1,2-glucanase from Talaromyces funiculosus with sophorose [Talaromyces funiculosus],6IMV_B The complex structure of endo-beta-1,2-glucanase from Talaromyces funiculosus with sophorose [Talaromyces funiculosus] |
|
| 1.24e-235 | 32 | 529 | 13 | 505 | The complex structure of endo-beta-1,2-glucanase mutant (E262Q) from Talaromyces funiculosus with beta-1,2-glucan [Talaromyces funiculosus],6IMW_B The complex structure of endo-beta-1,2-glucanase mutant (E262Q) from Talaromyces funiculosus with beta-1,2-glucan [Talaromyces funiculosus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
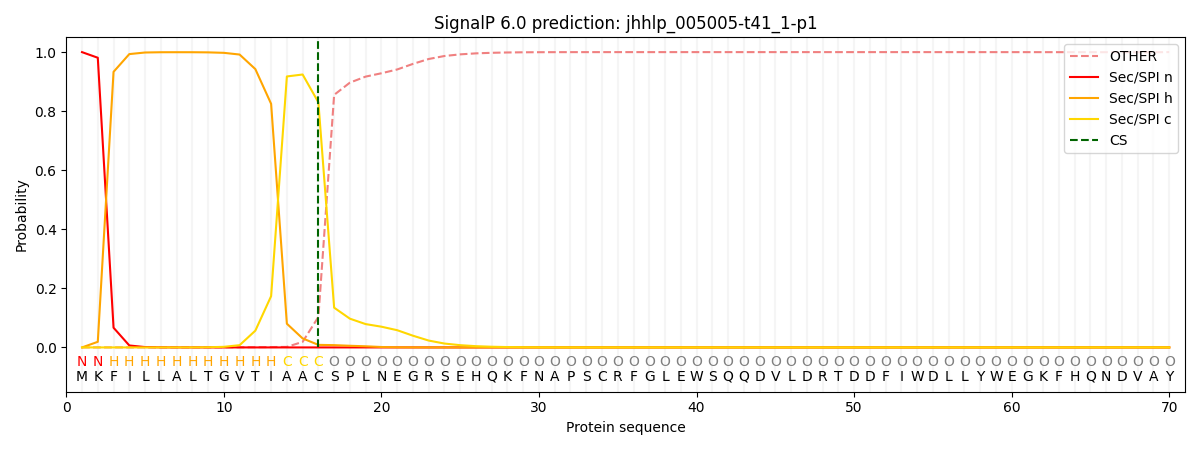
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000793 | 0.999180 | CS pos: 16-17. Pr: 0.8299 |
