You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_003835-t41_1-p1
Basic Information
help
| Species |
Lomentospora prolificans
|
| Lineage |
Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans
|
| CAZyme ID |
jhhlp_003835-t41_1-p1
|
| CAZy Family |
GH1 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 196 |
NLAX01000010|CGC17 |
21180.50 |
3.9076 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_LprolificansJHH5317 |
8768 |
N/A |
208 |
8560
|
|
| Gene Location |
No EC number prediction in jhhlp_003835-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA16 |
19 |
187 |
1.5e-67 |
0.9940119760479041 |
jhhlp_003835-t41_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.65e-83 |
4 |
191 |
5 |
193 |
| 3.65e-80 |
9 |
193 |
8 |
194 |
| 1.28e-79 |
9 |
193 |
57 |
243 |
| 1.95e-77 |
4 |
192 |
6 |
194 |
| 3.78e-75 |
5 |
193 |
7 |
195 |
jhhlp_003835-t41_1-p1 has no PDB hit.
jhhlp_003835-t41_1-p1 has no Swissprot hit.
This protein is predicted as SP
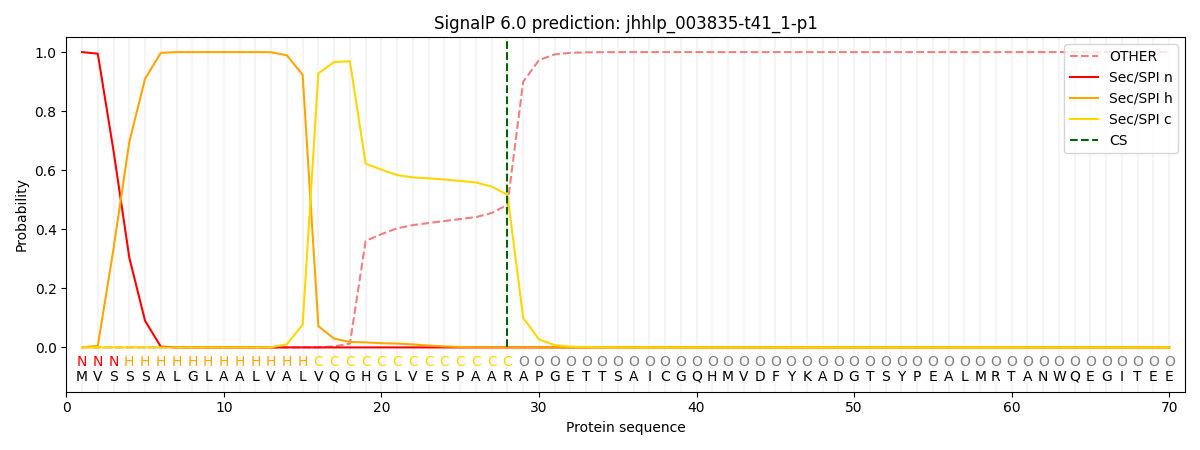
| Other |
SP_Sec_SPI |
CS Position |
| 0.000235 |
0.999749 |
CS pos: 28-29. Pr: 0.5170 |
There is no transmembrane helices in jhhlp_003835-t41_1-p1.

