You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_002658-t41_1-p1
You are here: Home > Sequence: jhhlp_002658-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_002658-t41_1-p1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 36 | 224 | 1e-21 | 0.7533039647577092 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 273828 | esterase_phb | 9.03e-40 | 41 | 236 | 3 | 194 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| 226040 | LpqC | 3.80e-24 | 26 | 282 | 36 | 277 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 402228 | Esterase_phd | 6.73e-21 | 37 | 231 | 1 | 190 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| 395257 | Peptidase_S9 | 7.27e-09 | 120 | 228 | 52 | 162 | Prolyl oligopeptidase family. |
| 224423 | DAP2 | 5.65e-08 | 37 | 166 | 378 | 507 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.28e-159 | 12 | 307 | 10 | 305 | |
| 6.06e-125 | 11 | 260 | 8 | 256 | |
| 2.75e-118 | 11 | 302 | 7 | 306 | |
| 6.45e-118 | 11 | 314 | 7 | 318 | |
| 6.45e-118 | 11 | 314 | 7 | 318 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.40e-77 | 20 | 292 | 1 | 272 | Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
|
| 5.44e-06 | 32 | 165 | 16 | 142 | C-terminal esterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.68e-88 | 19 | 292 | 31 | 303 | Probable acetylxylan esterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=axeA PE=3 SV=1 |
|
| 9.54e-88 | 19 | 292 | 31 | 303 | Acetylxylan esterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=axeA PE=1 SV=1 |
|
| 9.54e-88 | 19 | 292 | 31 | 303 | Probable acetylxylan esterase A OS=Aspergillus flavus OX=5059 GN=axeA PE=3 SV=1 |
|
| 6.98e-87 | 22 | 292 | 31 | 300 | Acetylxylan esterase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=axeA PE=1 SV=2 |
|
| 2.09e-80 | 19 | 292 | 28 | 300 | Acetylxylan esterase A OS=Aspergillus ficuum OX=5058 GN=axeA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
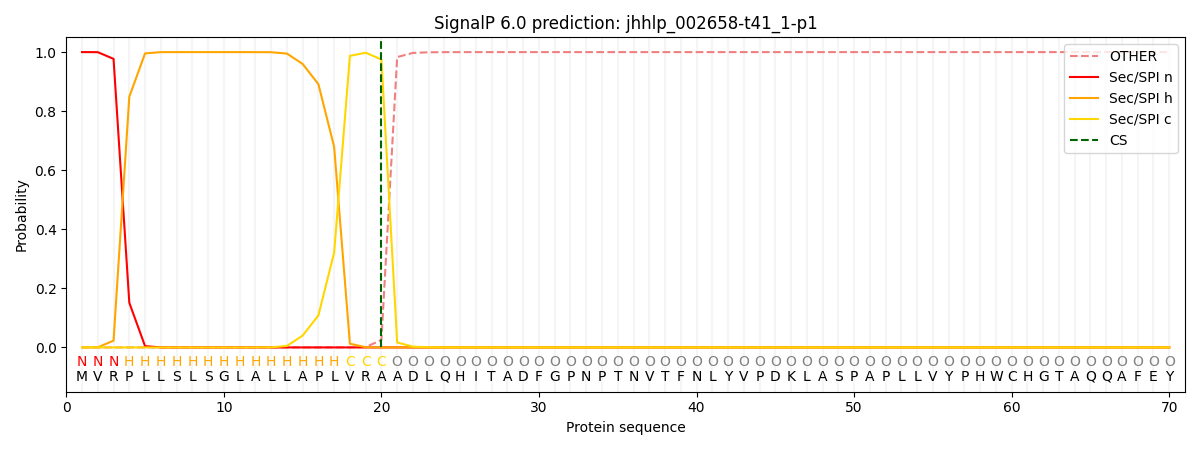
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000225 | 0.999773 | CS pos: 20-21. Pr: 0.9751 |
