You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_001710-t41_1-p1
You are here: Home > Sequence: jhhlp_001710-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_001710-t41_1-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 169878; End:174699 Strand: + | |||||||||||
Full Sequence Download help
| MTKLKTSSIP LLFLAGSSVA SPAPLAQDSV FFRVPESVSS ESVHNVHITY GDKLPQGNVN | 60 |
| IVYGDCASRD VGQSHHEVAT LFLSGSDSMP DRLVWSVPKD VTQRGCLHAF SESRLLGRSE | 120 |
| PVSFSESLQK RESLHEIGDS TGLWFNGVKY LKSTDKSTTV TDAAKSKKIA IIGGGISGLM | 180 |
| TSLLLTSVGI NDWHIIESTE RIGGRIRTKY LNGTSPSDYQ YQEMGPMRFP VSIRYSDTNE | 240 |
| TLDIQDHKMV FQLADVLNEM NGNSSDLAVN FIPWIQSNPN TPANSRGGRL PNGQIPSSAQ | 300 |
| IAADPSLRNP SATAADPEEA ELASEFVEHF LDFTPERMRN ISTNIFQAHR EAIDKGLFHW | 360 |
| SETAFLRYKL GLDADTVDLL SGSDNSPMWE EWYDNTYFGA TTWRTIDKGL NSLPRAFEPH | 420 |
| VQGKITYGRK VEGLAYDNAT SKVSVNWRDD EFAMVPQSEE YDYAMVAVPF SKVRLWRTPQ | 480 |
| YSSLLSRAIS TLNYAQSCKV ALHYKTRFWE HQENPIFGGC GSADIPGVSS ICYPSYQINS | 540 |
| TGPGVILGSY VSGTMARSVA ALSEKDHVAM IQRAMVEVHG EIANEEYTGN YNRQCWEVDE | 600 |
| HQAGAWASPT VGQQELFIPA YHNTEYNTIF IGEHTSITHA WIFSALESAV RGTTQLLLDL | 660 |
| GLVDEAKQVK PHSVRFSEDG FGDGPVQVSL PSWQWPGTYT YIDAFQDAGI QIRTDGGSNG | 720 |
| VNVGFNWNPS DIDPVNGTRS SSRKAYWDPA SERPNLSIIV NTFVSTVSFE NKTASGVNII | 780 |
| SRDTGDTVFV PASAEVILAA GSVHTPQILQ LSGIGPRDLL QELDIEEVVD LPGVGSNFQD | 840 |
| HLSISTAWRF TSEPEVNPSI LRDPDFLDAA FDEYWANKSG PITHTGKSCR VMLSLHNLTD | 900 |
| EVDSIVSTLE EEDPAQYLGD VYLASEELLA GYKAQHAILS RMVGSDEVSV FEHTFGGGAD | 960 |
| MSFHPQKPFS RGTIRIKSTD PHPADSPPQV DFRAMTHPLD MQIAILGLKF GRNILNGDIM | 1020 |
| GELGSREVSP GANVTSDDEL EQFLRTRAVG GLLGAEVFAA LQKEGVFTVS VLSLSSSKST | 1080 |
| FPSHIQVYKL DDDYPLDQLV GAFKGKNALV STLPGRPYAG HIRMVDVAVQ AGVKRFIPSE | 1140 |
| YGNNTCAGAA EQVDLYADKA KIITHLKSKE NTGLTWTAIH TGQFFDWGME SGWLNYCLDG | 1200 |
| RRVTIYNSGN KPWSTSTLGR QSPIKA | 1226 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 671 | 1048 | 2.3e-83 | 0.6390845070422535 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235000 | PRK02106 | 1.30e-47 | 678 | 1048 | 138 | 468 | choline dehydrogenase; Validated |
| 225186 | BetA | 7.19e-42 | 682 | 1048 | 147 | 469 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 396255 | Amino_oxidase | 7.54e-35 | 179 | 651 | 4 | 440 | Flavin containing amine oxidoreductase. This family consists of various amine oxidases, including maze polyamine oxidase (PAO) and various flavin containing monoamine oxidases (MAO). The aligned region includes the flavin binding site of these enzymes. The family also contains phytoene dehydrogenases and related enzymes. In vertebrates MAO plays an important role regulating the intracellular levels of amines via there oxidation; these include various neurotransmitters, neurotoxins and trace amines. In lower eukaryotes such as aspergillus and in bacteria the main role of amine oxidases is to provide a source of ammonium. PAOs in plants, bacteria and protozoa oxidase spermidine and spermine to an aminobutyral, diaminopropane and hydrogen peroxide and are involved in the catabolism of polyamines. Other members of this family include tryptophan 2-monooxygenase, putrescine oxidase, corticosteroid binding proteins and antibacterial glycoproteins. |
| 187569 | PCBER_SDR_a | 3.11e-31 | 1050 | 1218 | 8 | 178 | phenylcoumaran benzylic ether reductase (PCBER) like, atypical (a) SDRs. PCBER and pinoresinol-lariciresinol reductases are NADPH-dependent aromatic alcohol reductases, and are atypical members of the SDR family. Other proteins in this subgroup are identified as eugenol synthase. These proteins contain an N-terminus characteristic of NAD(P)-binding proteins and a small C-terminal domain presumed to be involved in substrate binding, but they do not have the conserved active site Tyr residue typically found in SDRs. Numerous other members have unknown functions. The glycine rich NADP-binding motif in this subgroup is of 2 forms: GXGXXG and G[GA]XGXXG; it tends to be atypical compared with the forms generally seen in classical or extended SDRs. The usual SDR active site tetrad is not present, but a critical active site Lys at the usual SDR position has been identified in various members, though other charged and polar residues are found at this position in this subgroup. Atypical SDR-related proteins retain the Rossmann fold of the SDRs, but have limited sequence identity and generally lack the catalytic properties of the archetypical members. Atypical SDRs include biliverdin IX beta reductase (BVR-B,aka flavin reductase), NMRa (a negative transcriptional regulator of various fungi), progesterone 5-beta-reductase like proteins, phenylcoumaran benzylic ether and pinoresinol-lariciresinol reductases, phenylpropene synthases, eugenol synthase, triphenylmethane reductase, isoflavone reductases, and others. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold, an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Sequence identity between different SDR enzymes is typically in the 15-30% range; they catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase numbering). In addition to the Tyr and Lys, there is often an upstream Ser and/or an Asn, contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. In addition to the Rossmann fold core region typical of all SDRs, extended SDRs have a less conserved C-terminal extension of approximately 100 amino acids, and typically have a TGXXGXXG cofactor binding motif. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. |
| 224152 | YobN | 2.63e-27 | 409 | 657 | 207 | 444 | Monoamine oxidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UQC77231.1|AA3_2 | 1.25e-83 | 669 | 1062 | 175 | 569 |
| UPK99906.1|AA3_2 | 3.35e-73 | 669 | 1045 | 172 | 546 |
| QSS75262.1|AA3_2 | 2.54e-69 | 669 | 1053 | 45 | 423 |
| SMR50915.1|AA3_2 | 3.25e-65 | 684 | 1043 | 191 | 548 |
| SMR51855.1|AA3_2 | 3.25e-65 | 684 | 1043 | 191 | 548 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZE2_A | 7.99e-45 | 684 | 1045 | 153 | 514 | Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE2_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE3_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE4_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE4_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE5_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE5_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE6_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE6_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE7_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE7_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],7AA2_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],7AA2_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495] |
| 6O9N_A | 4.55e-36 | 680 | 1048 | 191 | 573 | Structural insights on a new fungal aryl-alcohol oxidase [Thermothelomyces thermophilus],6O9N_B Structural insights on a new fungal aryl-alcohol oxidase [Thermothelomyces thermophilus] |
| 7C3I_A | 5.21e-24 | 167 | 657 | 8 | 492 | Structure of L-lysine oxidase D212A/D315A [Trichoderma viride],7C3I_B Structure of L-lysine oxidase D212A/D315A [Trichoderma viride],7C3J_A Structure of L-lysine oxidase D212A/D315A in complex with L-phenylalanine [Trichoderma viride],7C3J_B Structure of L-lysine oxidase D212A/D315A in complex with L-phenylalanine [Trichoderma viride],7C3L_A Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine [Trichoderma viride],7C3L_B Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine [Trichoderma viride] |
| 7C3H_A | 3.83e-23 | 167 | 657 | 8 | 492 | Structure of L-lysine oxidase in complex with L-lysine [Trichoderma viride],7C3H_B Structure of L-lysine oxidase in complex with L-lysine [Trichoderma viride] |
| 3X0V_A | 3.85e-23 | 167 | 657 | 8 | 492 | Structure of L-lysine oxidase [Trichoderma viride],3X0V_B Structure of L-lysine oxidase [Trichoderma viride] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P23623|OXLA_NEUCR | 3.09e-162 | 15 | 667 | 16 | 683 | L-amino-acid oxidase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=lox PE=1 SV=2 |
| sp|Q5AUN2|XPTC_EMENI | 3.20e-45 | 683 | 1045 | 184 | 545 | Dehydrogenase xptC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xptC PE=3 SV=1 |
| sp|Q0CJ60|ATC_ASPTN | 4.58e-33 | 684 | 1051 | 193 | 531 | Cyclase atC OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=atC PE=1 SV=1 |
| sp|A7MFA8|BETA_CROS8 | 1.32e-32 | 682 | 1065 | 141 | 492 | Oxygen-dependent choline dehydrogenase OS=Cronobacter sakazakii (strain ATCC BAA-894) OX=290339 GN=betA PE=3 SV=1 |
| sp|A5WA97|BETA_PSEP1 | 1.41e-32 | 682 | 1048 | 143 | 470 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas putida (strain ATCC 700007 / DSM 6899 / BCRC 17059 / F1) OX=351746 GN=betA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
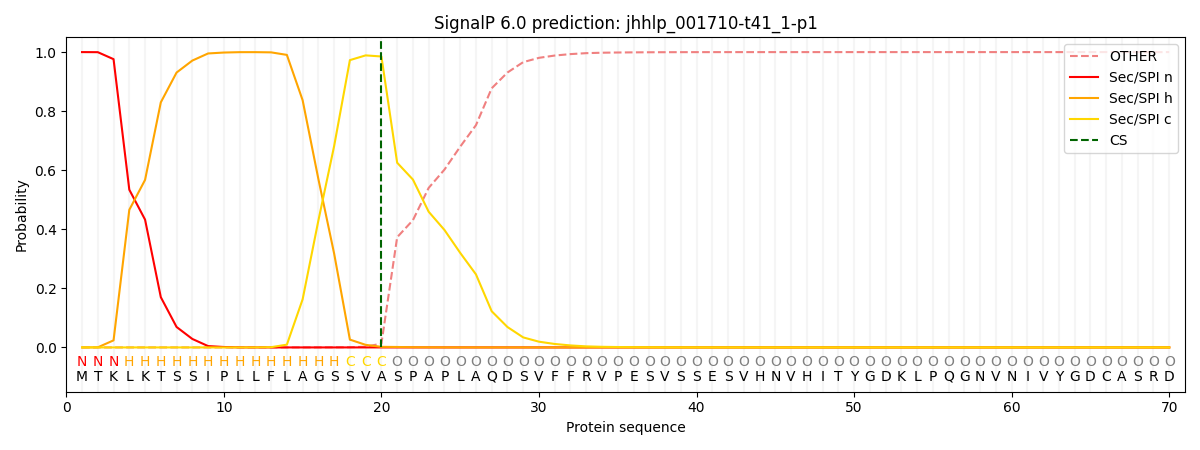
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000438 | 0.999556 | CS pos: 20-21. Pr: 0.9852 |
