You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_000986-t41_1-p1
Basic Information
help
| Species |
Lomentospora prolificans
|
| Lineage |
Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans
|
| CAZyme ID |
jhhlp_000986-t41_1-p1
|
| CAZy Family |
AA4 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 642 |
|
72516.18 |
6.3418 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_LprolificansJHH5317 |
8768 |
N/A |
208 |
8560
|
|
| Gene Location |
No EC number prediction in jhhlp_000986-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH76 |
158 |
494 |
3e-50 |
0.6452513966480447 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 397638
|
Glyco_hydro_76 |
3.37e-13 |
231 |
499 |
126 |
339 |
Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.91e-193 |
1 |
610 |
1 |
614 |
| 1.91e-193 |
1 |
610 |
1 |
614 |
| 1.91e-193 |
1 |
610 |
1 |
614 |
| 7.69e-193 |
1 |
610 |
1 |
614 |
| 1.44e-191 |
1 |
610 |
1 |
608 |
jhhlp_000986-t41_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.64e-06 |
233 |
463 |
164 |
354 |
Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
This protein is predicted as SP
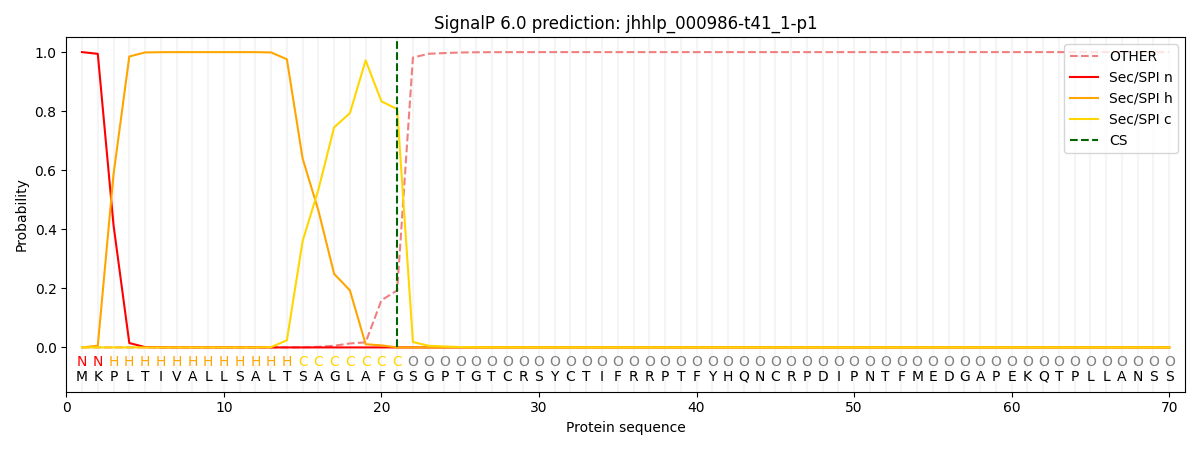
| Other |
SP_Sec_SPI |
CS Position |
| 0.000222 |
0.999740 |
CS pos: 21-22. Pr: 0.8068 |
There is no transmembrane helices in jhhlp_000986-t41_1-p1.

