You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_000934-t41_1-p1
Basic Information
help
| Species |
Lomentospora prolificans
|
| Lineage |
Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans
|
| CAZyme ID |
jhhlp_000934-t41_1-p1
|
| CAZy Family |
AA3 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 846 |
|
94710.66 |
7.6774 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_LprolificansJHH5317 |
8768 |
N/A |
208 |
8560
|
|
| Gene Location |
No EC number prediction in jhhlp_000934-t41_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT90 |
524 |
832 |
1.1e-45 |
0.98 |
jhhlp_000934-t41_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 2.18e-168 |
18 |
845 |
80 |
966 |
| 4.98e-168 |
18 |
841 |
95 |
964 |
| 5.49e-166 |
18 |
841 |
91 |
973 |
| 1.86e-163 |
18 |
841 |
101 |
987 |
| 5.98e-163 |
18 |
842 |
76 |
960 |
jhhlp_000934-t41_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.49e-38 |
305 |
845 |
152 |
669 |
Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
CS Position |
| 0.999346 |
0.000656 |
|
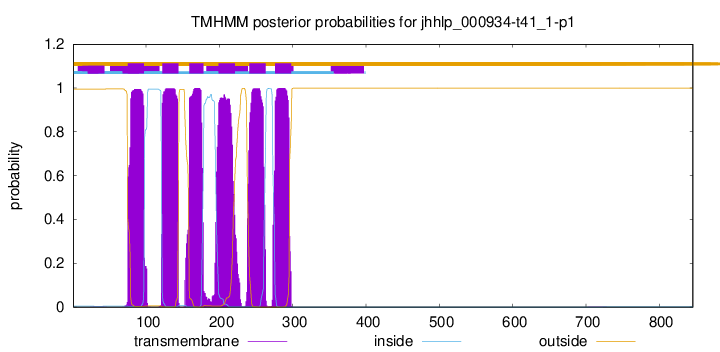
| Start |
End |
| 75 |
97 |
| 122 |
144 |
| 159 |
178 |
| 199 |
221 |
| 241 |
263 |
| 276 |
298 |


