You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_000275-t41_1-p1
You are here: Home > Sequence: jhhlp_000275-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_000275-t41_1-p1 | |||||||||||
| CAZy Family | AA16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 32 | 422 | 1.1e-165 | 0.9825436408977556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 3.34e-22 | 8 | 426 | 23 | 398 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| 271333 | NHL_like_5 | 0.002 | 47 | 116 | 189 | 256 | Uncharacterized NHL-repeat domain in bacterial proteins. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
| 271327 | NHL_like_2 | 0.009 | 41 | 150 | 95 | 189 | Uncharacterized NHL-repeat domain in bacterial and archaeal proteins. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.48e-165 | 5 | 433 | 3 | 440 | |
| 6.95e-164 | 21 | 428 | 122 | 515 | |
| 5.46e-162 | 17 | 458 | 23 | 473 | |
| 3.56e-161 | 23 | 434 | 22 | 437 | |
| 1.82e-157 | 9 | 443 | 6 | 448 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.25e-148 | 29 | 425 | 3 | 403 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.29e-148 | 29 | 425 | 4 | 404 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 1.33e-148 | 29 | 425 | 5 | 405 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 2.62e-71 | 25 | 424 | 4 | 410 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.62e-17 | 36 | 425 | 71 | 444 | L-sorbosone dehydrogenase OS=Gluconacetobacter liquefaciens OX=89584 PE=4 SV=1 |
|
| 1.53e-09 | 7 | 422 | 10 | 374 | Uncharacterized protein BB_0024 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0024 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
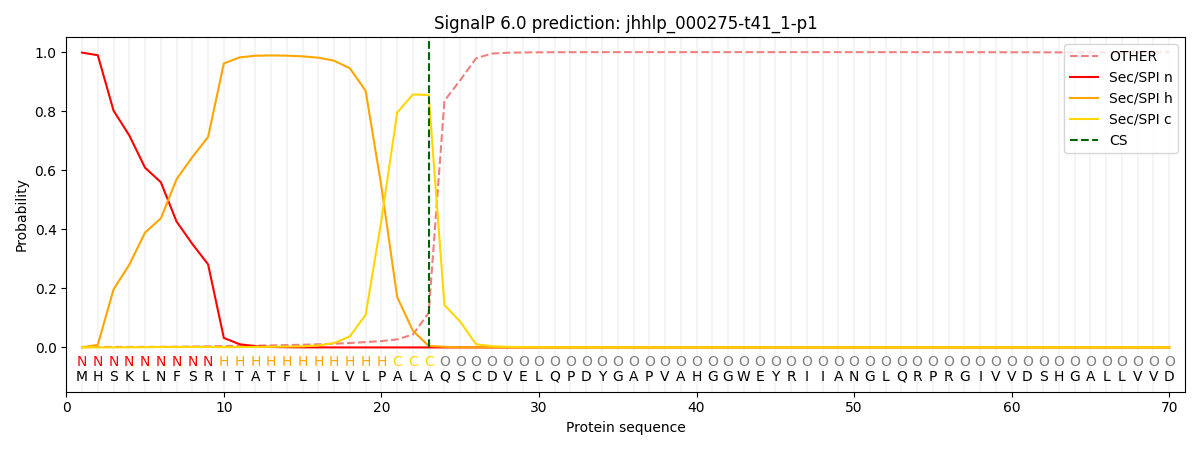
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002835 | 0.997138 | CS pos: 23-24. Pr: 0.8548 |
