You are browsing environment: FUNGIDB
CAZyme Information: jhhlp_000268-t41_1-p1
You are here: Home > Sequence: jhhlp_000268-t41_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lomentospora prolificans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Microascaceae; Lomentospora; Lomentospora prolificans | |||||||||||
| CAZyme ID | jhhlp_000268-t41_1-p1 | |||||||||||
| CAZy Family | AA13|CBM20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 213 | 750 | 2.7e-71 | 0.9762773722627737 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 366272 | GMC_oxred_N | 6.12e-55 | 268 | 497 | 2 | 218 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 225186 | BetA | 5.40e-43 | 221 | 742 | 9 | 526 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 398739 | GMC_oxred_C | 6.02e-29 | 576 | 745 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 235000 | PRK02106 | 3.41e-09 | 664 | 748 | 457 | 534 | choline dehydrogenase; Validated |
| 223717 | FixC | 5.75e-08 | 217 | 251 | 1 | 35 | Dehydrogenase (flavoprotein) [Energy production and conversion]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.62e-206 | 14 | 753 | 16 | 734 | |
| 1.95e-15 | 219 | 753 | 13 | 602 | |
| 6.01e-15 | 214 | 754 | 8 | 603 | |
| 1.81e-14 | 214 | 753 | 8 | 602 | |
| 5.50e-14 | 214 | 753 | 8 | 602 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.12e-08 | 217 | 754 | 3 | 569 | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase [Aspergillus flavus NRRL3357],4YNU_A Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone [Aspergillus flavus NRRL3357] |
|
| 6.10e-08 | 218 | 750 | 228 | 764 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
|
| 1.76e-06 | 661 | 753 | 412 | 512 | Chain A, 6'''-hydroxyparomomycin C oxidase [Microbacterium trichothecenolyticum],7DVE_B Chain B, 6'''-hydroxyparomomycin C oxidase [Microbacterium trichothecenolyticum] |
|
| 3.43e-06 | 219 | 753 | 6 | 636 | Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_B Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_C Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_D Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_E Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_F Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_G Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_H Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5I68_A Chain A, Alcohol oxidase 1 [Komagataella pastoris] |
|
| 7.01e-06 | 221 | 750 | 7 | 528 | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state [Methylovorus sp. MP688],4UDP_B Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state [Methylovorus sp. MP688],4UDQ_A Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state [Methylovorus sp. MP688],4UDQ_B Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state [Methylovorus sp. MP688] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.22e-84 | 122 | 754 | 103 | 747 | Long-chain-alcohol oxidase FAO2 OS=Lotus japonicus OX=34305 GN=FAO2 PE=2 SV=1 |
|
| 1.19e-80 | 218 | 754 | 235 | 745 | Long-chain-alcohol oxidase FAO1 OS=Lotus japonicus OX=34305 GN=FAO1 PE=1 SV=1 |
|
| 3.95e-75 | 218 | 754 | 236 | 743 | Long-chain-alcohol oxidase FAO4B OS=Arabidopsis thaliana OX=3702 GN=FAO4B PE=2 SV=2 |
|
| 9.61e-75 | 112 | 754 | 57 | 723 | Long-chain-alcohol oxidase FAO4A OS=Arabidopsis thaliana OX=3702 GN=FAO4A PE=3 SV=2 |
|
| 2.83e-70 | 218 | 754 | 244 | 753 | Long-chain-alcohol oxidase FAO1 OS=Arabidopsis thaliana OX=3702 GN=FAO1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
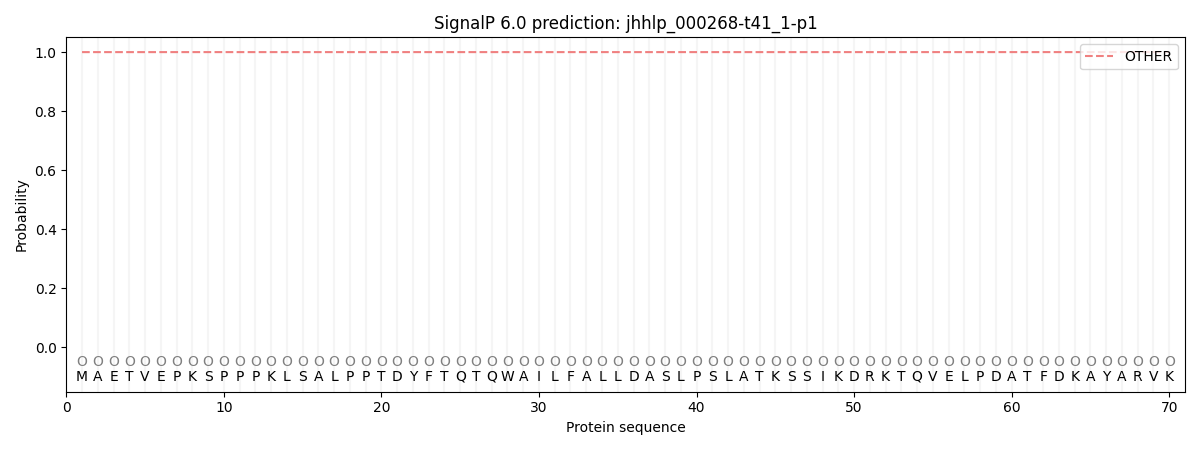
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999977 | 0.000051 |
