You are browsing environment: FUNGIDB
CAZyme Information: evm.model.AC2VRR_s00012g53-p1
You are here: Home > Sequence: evm.model.AC2VRR_s00012g53-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
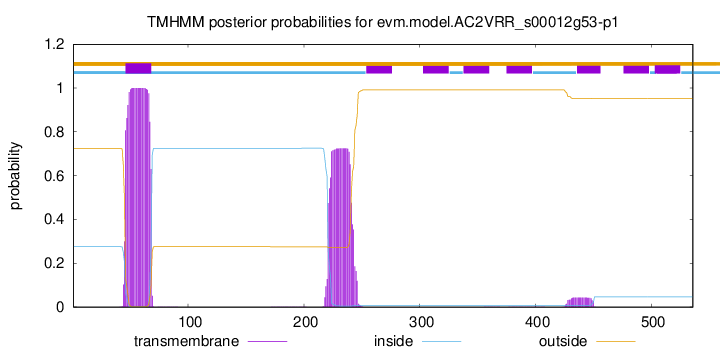
TMHMM annotations
Basic Information help
| Species | Albugo candida | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Albuginaceae; Albugo; Albugo candida | |||||||||||
| CAZyme ID | evm.model.AC2VRR_s00012g53-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 140 | 488 | 4.3e-137 | 0.997093023255814 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 536 | 1 | 536 | |
| 1.31e-183 | 7 | 535 | 11 | 540 | |
| 1.17e-94 | 34 | 535 | 25 | 520 | |
| 7.22e-93 | 258 | 535 | 3 | 278 | |
| 9.03e-93 | 6 | 535 | 103 | 627 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.17e-16 | 137 | 510 | 22 | 361 | The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
|
| 1.56e-16 | 137 | 510 | 22 | 361 | Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
|
| 1.97e-16 | 137 | 510 | 55 | 394 | Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_A Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_B Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_C Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3] |
|
| 1.97e-16 | 137 | 510 | 55 | 394 | Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHO_B Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHO_C Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii] |
|
| 3.49e-16 | 137 | 510 | 55 | 394 | Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHM_B Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHM_C Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.94e-18 | 124 | 516 | 39 | 373 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
|
| 1.47e-14 | 142 | 521 | 41 | 368 | Major extracellular endoglucanase OS=Xanthomonas campestris pv. campestris (strain ATCC 33913 / DSM 3586 / NCPPB 528 / LMG 568 / P 25) OX=190485 GN=engXCA PE=1 SV=2 |
|
| 3.39e-08 | 137 | 511 | 57 | 399 | Endoglucanase D OS=Cellulomonas fimi OX=1708 GN=cenD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999893 | 0.000128 |