You are browsing environment: FUNGIDB
CAZyme Information: evm.model.AC2VRR_s00008g102-p1
You are here: Home > Sequence: evm.model.AC2VRR_s00008g102-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Albugo candida | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Albuginaceae; Albugo; Albugo candida | |||||||||||
| CAZyme ID | evm.model.AC2VRR_s00008g102-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 849 | 1542 | 1.3e-264 | 0.8958051420838972 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 1.20e-173 | 850 | 1532 | 3 | 700 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 2.29e-32 | 128 | 223 | 5 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 273317 | SP | 2.55e-08 | 1843 | 2199 | 100 | 469 | MFS transporter, sugar porter (SP) family. This model represent the sugar porter subfamily of the major facilitator superfamily (pfam00083) [Transport and binding proteins, Carbohydrates, organic alcohols, and acids] |
| 395036 | Sugar_tr | 1.69e-07 | 1843 | 2199 | 75 | 436 | Sugar (and other) transporter. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 2277 | 1 | 2275 | |
| 0.0 | 24 | 2277 | 10 | 2320 | |
| 0.0 | 52 | 2282 | 98 | 2299 | |
| 0.0 | 19 | 2220 | 12 | 2230 | |
| 0.0 | 17 | 2265 | 12 | 2225 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.49e-231 | 2 | 1755 | 178 | 1931 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
|
| 8.84e-230 | 16 | 1755 | 193 | 1931 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
|
| 4.87e-229 | 50 | 1755 | 239 | 1910 | Callose synthase 5 OS=Arabidopsis thaliana OX=3702 GN=CALS5 PE=1 SV=1 |
|
| 5.90e-225 | 2 | 1755 | 182 | 1936 | Callose synthase 3 OS=Arabidopsis thaliana OX=3702 GN=CALS3 PE=3 SV=3 |
|
| 4.80e-219 | 49 | 1762 | 254 | 1922 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000028 | 0.000000 |
TMHMM Annotations download full data without filtering help
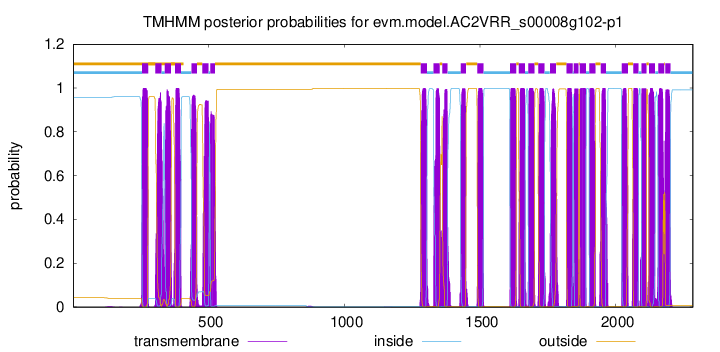
| Start | End |
|---|---|
| 1429 | 1448 |
| 1490 | 1512 |
| 1611 | 1633 |
| 1643 | 1665 |
| 1678 | 1700 |
| 1715 | 1737 |
| 1758 | 1780 |
| 1819 | 1841 |
| 1846 | 1863 |
| 1868 | 1890 |
| 1903 | 1925 |
| 1945 | 1964 |
| 2023 | 2045 |
| 2065 | 2084 |
| 2096 | 2113 |
| 2123 | 2145 |
| 2157 | 2179 |
| 2183 | 2202 |
| 254 | 276 |
| 303 | 325 |
| 338 | 360 |
| 375 | 397 |
| 436 | 456 |
| 476 | 498 |
| 505 | 522 |
| 1282 | 1304 |
| 1329 | 1351 |
| 1361 | 1380 |
