You are browsing environment: FUNGIDB
CAZyme Information: ZTRI_8.359.mRNA-p1
You are here: Home > Sequence: ZTRI_8.359.mRNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Zymoseptoria tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici | |||||||||||
| CAZyme ID | ZTRI_8.359.mRNA-p1 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 5 | 394 | 7.7e-77 | 0.9897172236503856 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178772 | PLN03233 | 0.0 | 624 | 1137 | 12 | 510 | putative glutamate-tRNA ligase; Provisional |
| 215492 | PLN02907 | 0.0 | 506 | 1137 | 73 | 717 | glutamate-tRNA ligase |
| 240404 | PTZ00402 | 6.45e-175 | 617 | 1136 | 45 | 552 | glutamyl-tRNA synthetase; Provisional |
| 273091 | gltX_arch | 8.49e-167 | 604 | 1133 | 73 | 556 | glutamyl-tRNA synthetase, archaeal and eukaryotic family. The glutamyl-tRNA synthetases of the eukaryotic cytosol and of the Archaea are more similar to glutaminyl-tRNA synthetases than to bacterial glutamyl-tRNA synthetases. This model models just the eukaryotic cytosolic and archaeal forms of the enzyme. In some eukaryotes, the glutamyl-tRNA synthetase is part of a longer, multifunctional aminoacyl-tRNA ligase. In many species, the charging of tRNA(gln) proceeds first through misacylation with Glu and then transamidation. For this reason, glutamyl-tRNA synthetases, including all known archaeal enzymes (as of 2010) may act on both tRNA(gln) and tRNA(glu). [Protein synthesis, tRNA aminoacylation] |
| 235424 | PRK05347 | 1.63e-133 | 622 | 1126 | 28 | 539 | glutaminyl-tRNA synthetase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1138 | 1 | 1138 | |
| 0.0 | 1 | 1138 | 1 | 1138 | |
| 0.0 | 1 | 479 | 1 | 479 | |
| 0.0 | 1 | 479 | 1 | 479 | |
| 6.87e-261 | 1 | 479 | 1 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.60e-86 | 624 | 1128 | 28 | 538 | Chain A, Glutaminyl-tRNA synthetase [Escherichia coli K-12],2RE8_A Chain A, Glutaminyl-tRNA synthetase [Escherichia coli] |
|
| 3.83e-85 | 624 | 1128 | 20 | 530 | Unliganded glutaminyl-tRNA synthetase [Escherichia coli] |
|
| 4.87e-85 | 624 | 1128 | 28 | 538 | Crystal Structure Of Glutaminyl-Trna Synthetase Complexed With A Trna- Gln Mutant And An Active-Site Inhibitor [Escherichia coli],1EUY_A GLUTAMINYL-TRNA SYNTHETASE COMPLEXED WITH A TRNA MUTANT AND AN ACTIVE SITE INHIBITOR [Escherichia coli],1EXD_A Crystal Structure Of A Tight-Binding Glutamine Trna Bound To Glutamine Aminoacyl Trna Synthetase [Escherichia coli] |
|
| 5.56e-85 | 624 | 1128 | 27 | 537 | Structure of E.coli glutaminyl-tRNA synthetase complexed with trnagln and ATP at 2.8 Angstroms resolution [Escherichia coli],1GTR_A Structural Basis Of Anticodon Loop Recognition By Glutaminyl-Trna Synthetase [Escherichia coli],1GTS_A Structural Basis For Transfer Rna Aminoaceylation By Escherichia Coli Glutaminyl-trna Synthetase [Escherichia coli],1QTQ_A Glutaminyl-Trna Synthetase Complexed With Trna And An Amino Acid Analog [Escherichia coli],1ZJW_A Glutaminyl-tRNA synthetase complexed to glutamine and 2'deoxy A76 glutamine tRNA [Escherichia coli],4JXX_A Crystal structure of E coli E. coli glutaminyl-tRNA synthetase bound to tRNA(Gln)(CUG) and ATP from novel cryostabilization conditions [Escherichia coli K-12],4JXZ_A Structure of E. coli glutaminyl-tRNA synthetase bound to ATP and a tRNA(Gln) acceptor containing a UUG anticodon [Escherichia coli K-12],4JYZ_A Crystal structure of E. coli glutaminyl-tRNA synthetase bound to ATP and native tRNA(Gln) containing the cmnm5s2U34 anticodon wobble base [Escherichia coli K-12] |
|
| 5.70e-85 | 624 | 1128 | 28 | 538 | Chain A, Glutaminyl-tRNA synthetase [Escherichia coli],1O0C_A Chain A, Glutaminyl-tRNA synthetase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.84e-226 | 512 | 1136 | 88 | 707 | Probable glutamate--tRNA ligase, cytoplasmic OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gus1 PE=1 SV=1 |
|
| 2.97e-213 | 506 | 1138 | 69 | 701 | Glutamate--tRNA ligase, cytoplasmic OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GUS1 PE=1 SV=3 |
|
| 1.70e-181 | 510 | 1136 | 79 | 716 | Glutamate--tRNA ligase, cytoplasmic OS=Arabidopsis thaliana OX=3702 GN=At5g26710 PE=1 SV=1 |
|
| 1.29e-171 | 1 | 479 | 1 | 492 | GPI mannosyltransferase 4 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=SMP3 PE=3 SV=2 |
|
| 3.15e-166 | 469 | 1135 | 42 | 727 | Probable glutamate--tRNA ligase, cytoplasmic OS=Dictyostelium discoideum OX=44689 GN=gluS PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.249914 | 0.750076 | CS pos: 23-24. Pr: 0.5134 |
TMHMM Annotations download full data without filtering help
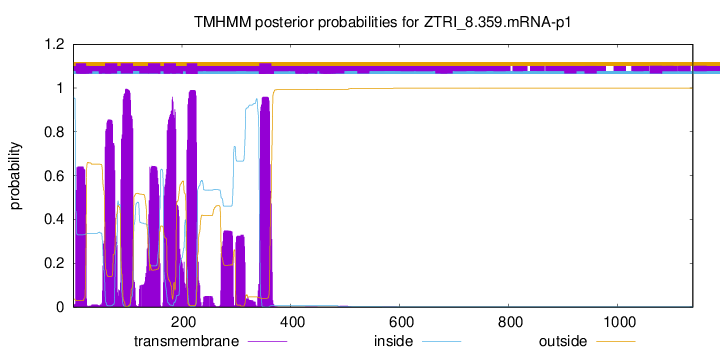
| Start | End |
|---|---|
| 5 | 24 |
| 59 | 81 |
| 88 | 110 |
| 138 | 160 |
| 167 | 189 |
| 209 | 228 |
| 342 | 364 |
