You are browsing environment: FUNGIDB
CAZyme Information: ZTRI_1.1606.mRNA-p1
You are here: Home > Sequence: ZTRI_1.1606.mRNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
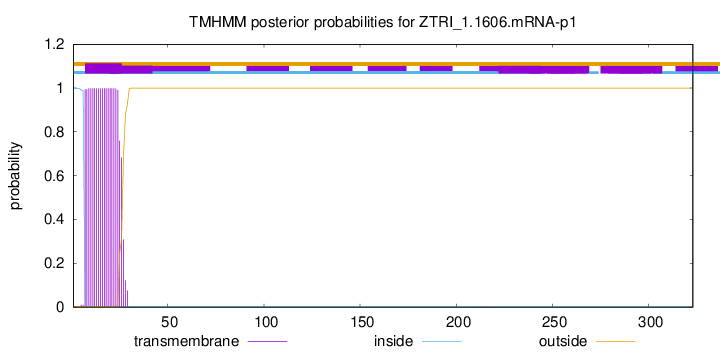
TMHMM annotations
Basic Information help
| Species | Zymoseptoria tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici | |||||||||||
| CAZyme ID | ZTRI_1.1606.mRNA-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT64 | 73 | 314 | 2.2e-56 | 0.9717741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 401264 | Glyco_transf_64 | 1.20e-56 | 73 | 311 | 1 | 235 | Glycosyl transferase family 64 domain. Members of this family catalyze the transfer reaction of N-acetylglucosamine and N-acetylgalactosamine from the respective UDP-sugars to the non-reducing end of [glucuronic acid]beta 1-3[galactose]beta 1-O-naphthalenemethanol, an acceptor substrate analog of the natural common linker of various glycosylaminoglycans. They are also required for the biosynthesis of heparan-sulphate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.11e-238 | 1 | 323 | 1 | 323 | |
| 5.11e-238 | 1 | 323 | 1 | 323 | |
| 7.02e-217 | 1 | 323 | 1 | 301 | |
| 2.86e-216 | 1 | 323 | 1 | 301 | |
| 7.85e-41 | 69 | 317 | 87 | 337 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.91e-28 | 44 | 304 | 602 | 859 | Chain A, Exostosin-like 3 [Homo sapiens],7AU2_B Chain B, Exostosin-like 3 [Homo sapiens],7AUA_A Chain A, Exostosin-like 3 [Homo sapiens],7AUA_B Chain B, Exostosin-like 3 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.34e-31 | 72 | 319 | 73 | 331 | Glycosylinositol phosphorylceramide mannosyl transferase 1 OS=Arabidopsis thaliana OX=3702 GN=GMT1 PE=1 SV=1 |
|
| 1.53e-30 | 73 | 304 | 499 | 727 | Glucosamine inositolphosphorylceramide transferase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=GINT1 PE=2 SV=1 |
|
| 1.53e-30 | 73 | 304 | 499 | 727 | Glucosamine inositolphosphorylceramide transferase 1 OS=Oryza sativa subsp. indica OX=39946 GN=GINT1 PE=3 SV=1 |
|
| 6.02e-28 | 71 | 298 | 64 | 299 | Exostosin-like 2 OS=Homo sapiens OX=9606 GN=EXTL2 PE=1 SV=1 |
|
| 6.47e-28 | 65 | 304 | 448 | 685 | Exostosin-2 OS=Mus musculus OX=10090 GN=Ext2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.730474 | 0.269544 |