You are browsing environment: FUNGIDB
CAZyme Information: ZTRI_1.141.mRNA-p1
You are here: Home > Sequence: ZTRI_1.141.mRNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
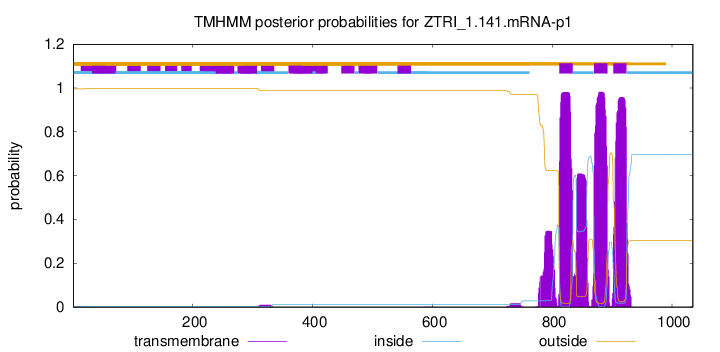
TMHMM annotations
Basic Information help
| Species | Zymoseptoria tritici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici | |||||||||||
| CAZyme ID | ZTRI_1.141.mRNA-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 425 | 625 | 7.4e-48 | 0.9948453608247423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 1.35e-37 | 425 | 625 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 395036 | Sugar_tr | 4.51e-36 | 793 | 994 | 59 | 256 | Sugar (and other) transporter. |
| 340914 | MFS_HXT | 6.99e-28 | 793 | 930 | 58 | 195 | Fungal Hexose transporter subfamily of the Major Facilitator Superfamily of transporters and similar proteins. The fungal hexose transporter (HXT) subfamily is comprised of functionally redundant proteins that function mainly in the transport of glucose, as well as other sugars such as galactose and fructose. Saccharomyces cerevisiae has 20 genes that encode proteins in this family (HXT1 to HXT17, GAL2, SNF3, and RGT2). Seven of these (HXT1-7) encode functional glucose transporters. Gal2p is a galactose transporter, while Rgt2p and Snf3p act as cell surface glucose receptors that initiate signal transduction in response to glucose, functioning in an induction pathway responsible for glucose uptake. Rgt2p is activated by high levels of glucose and stimulates expression of low affinity glucose transporters such as Hxt1p and Hxt3p, while Snf3p generates a glucose signal in response to low levels of glucose, stimulating the expression of high affinity glucose transporters such as Hxt2p and Hxt4p. Schizosaccharomyces pombe contains eight GHT genes (GHT1-8) belonging to this family. Ght1, Ght2, and Ght5 are high-affinity glucose transporters; Ght3 is a high-affinity gluconate transporter; and Ght6 high-affinity fructose transporter. The substrate specificities for Ght4, Ght7, and Ght8 remain undetermined. The HXT subfamily belongs to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 370992 | GPI-anchored | 3.46e-27 | 35 | 130 | 1 | 93 | Ser-Thr-rich glycosyl-phosphatidyl-inositol-anchored membrane family. Some members of this family appear to be serine- threonine-rich membrane-anchored proteins, anchored by glycosyl-phosphatidylinositol. In A. fumigatus these proteins play a role in fungal cell wall organisation. In Lentinula edodes this family is involved in fruiting body formation, and may have a more general role in signalling in other organisms as it interacts with MAPK. The family is also found in archaea and bacteria. |
| 340918 | MFS_HMIT_like | 1.08e-24 | 794 | 928 | 49 | 180 | H(+)-myo-inositol cotransporter and similar transporters of the Major Facilitator Superfamily. This subfamily is composed of myo-inositol/inositol transporters and similar transporters from vertebrates, plant, and fungi. The human protein is called H(+)-myo-inositol cotransporter/Proton myo-inositol cotransporter (HMIT), or H(+)-myo-inositol symporter, or Solute carrier family 2 member 13 (SLC2A13). HMIT is classified as a Class 3 GLUT (glucose transporter) based on sequence similarity with GLUTs, but it does not transport glucose. It specifically transports myo-inositol and is expressed predominantly in the brain, with high expression in the hippocampus, hypothalamus, cerebellum and brainstem. HMIT may be involved in regulating processes that require high levels of myo-inositol or its phosphorylated derivatives, such as membrane recycling, growth cone dynamics, and synaptic vesicle exocytosis. Arabidopsis Inositol transporter 4 (AtINT4) mediates high-affinity H+ symport of myo-inositol across the plasma membrane. The HMIT-like subfamily belongs to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 333 | 1009 | 31 | 707 | |
| 3.97e-235 | 333 | 692 | 31 | 390 | |
| 1.81e-233 | 333 | 692 | 31 | 390 | |
| 1.12e-229 | 333 | 692 | 31 | 382 | |
| 6.43e-45 | 425 | 633 | 7 | 209 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.19e-15 | 793 | 974 | 67 | 266 | Crystal structure of D-xylose-proton symporter [Escherichia coli K-12] |
|
| 1.22e-15 | 793 | 974 | 68 | 267 | Partially occluded inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA3_B Partially occluded inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA4_A Inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA4_B Inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA4_C Inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12] |
|
| 1.26e-15 | 793 | 974 | 68 | 267 | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose [Escherichia coli K-12],4GBZ_A The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose [Escherichia coli K-12],4GC0_A The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose [Escherichia coli K-12] |
|
| 1.40e-15 | 793 | 974 | 95 | 294 | Crystal structure of a double Trp XylE mutants (G58W/L315W) [Escherichia coli] |
|
| 2.02e-10 | 775 | 976 | 54 | 255 | Structure of the human glucose transporter GLUT3 / SLC2A3 [Homo sapiens],5C65_B Structure of the human glucose transporter GLUT3 / SLC2A3 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.86e-33 | 793 | 980 | 76 | 261 | MFS-type transporter oryC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=oryC PE=3 SV=1 |
|
| 1.17e-30 | 793 | 994 | 80 | 283 | Major facilitator superfamily transporter mfsA OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mfsA PE=2 SV=1 |
|
| 1.99e-25 | 793 | 973 | 90 | 273 | Sugar transporter STL1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=STL1 PE=1 SV=2 |
|
| 2.48e-17 | 793 | 996 | 132 | 330 | Galactose transporter OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAL2 PE=1 SV=3 |
|
| 9.59e-17 | 795 | 958 | 156 | 319 | Low glucose sensor SNF3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SNF3 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000200 | 0.999756 | CS pos: 20-21. Pr: 0.9667 |