You are browsing environment: FUNGIDB
CAZyme Information: YALI0_A11077g-t26_1-p1
You are here: Home > Sequence: YALI0_A11077g-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Yarrowia lipolytica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Dipodascaceae; Yarrowia; Yarrowia lipolytica | |||||||||||
| CAZyme ID | YALI0_A11077g-t26_1-p1 | |||||||||||
| CAZy Family | GT66 | |||||||||||
| CAZyme Description | YALI0A11077p | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.00e-28 | 28 | 284 | 45 | 299 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 239303 | PDI_a_ERp46 | 0.008 | 40 | 76 | 29 | 65 | PDIa family, endoplasmic reticulum protein 46 (ERp46) subfamily; ERp46 is an ER-resident protein containing three redox active TRX domains. Yeast complementation studies show that ERp46 can substitute for protein disulfide isomerase (PDI) function in vivo. It has been detected in many tissues, however, transcript and protein levels do not correlate in all tissues, suggesting regulation at a posttranscriptional level. An identical protein, named endoPDI, has been identified as an endothelial PDI that is highly expressed in the endothelium of tumors and hypoxic lesions. It has a protective effect on cells exposed to hypoxia. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.11e-219 | 1 | 294 | 1 | 294 | |
| 4.11e-218 | 1 | 294 | 1 | 294 | |
| 4.11e-218 | 1 | 294 | 1 | 294 | |
| 1.25e-47 | 24 | 290 | 53 | 319 | |
| 7.01e-47 | 24 | 290 | 125 | 391 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.58e-40 | 28 | 285 | 281 | 537 | Probable family 17 glucosidase SCW11 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW11 PE=1 SV=1 |
|
| 1.25e-24 | 20 | 284 | 287 | 553 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
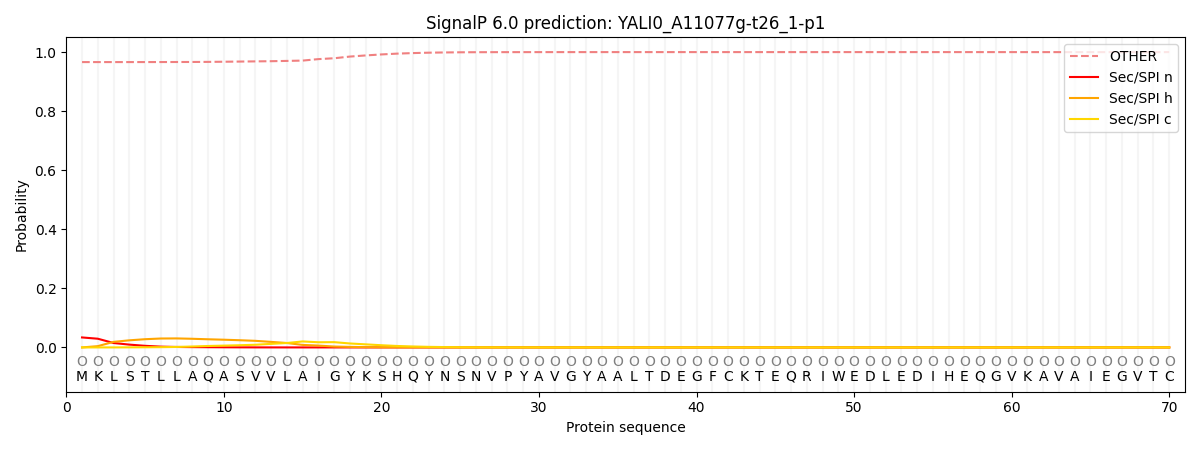
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.969091 | 0.030927 |
