You are browsing environment: FUNGIDB
CAZyme Information: XP_001912276.1
You are here: Home > Sequence: XP_001912276.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001912276.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Cutinase [Source:UniProtKB/TrEMBL;Acc:B2A9K8] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 62 | 241 | 2.2e-36 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 2.24e-56 | 62 | 242 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.40e-172 | 1 | 245 | 1 | 245 | |
| 2.40e-172 | 1 | 245 | 1 | 245 | |
| 1.63e-167 | 1 | 245 | 1 | 242 | |
| 4.37e-99 | 1 | 245 | 1 | 236 | |
| 4.37e-99 | 1 | 245 | 1 | 236 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.11e-75 | 51 | 242 | 3 | 193 | Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY3_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9. [Paraphoma sp. B47-9],7CY9_A Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9],7CY9_B Crystal structure of a biodegradable plastic-degrading cutinase from Paraphoma sp. B47-9 solved by getting the phase from anomalous scattering of uncovalently coordinated arsenic (cacodylate). [Paraphoma sp. B47-9] |
|
| 1.88e-69 | 53 | 244 | 19 | 209 | Chain A, CUTINASE [Fusarium vanettenii],1CUW_B Chain B, CUTINASE [Fusarium vanettenii] |
|
| 1.11e-68 | 36 | 245 | 6 | 212 | Crystal structure of Fusarium oxysporum cutinase [Fusarium oxysporum f. sp. raphani 54005],5AJH_B Crystal structure of Fusarium oxysporum cutinase [Fusarium oxysporum f. sp. raphani 54005],5AJH_C Crystal structure of Fusarium oxysporum cutinase [Fusarium oxysporum f. sp. raphani 54005] |
|
| 1.52e-68 | 39 | 244 | 8 | 209 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 2.59e-68 | 53 | 244 | 5 | 195 | CUTINASE, A LIPOLYTIC ENZYME WITH A PREFORMED OXYANION HOLE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.10e-70 | 1 | 244 | 1 | 226 | Cutinase 2 OS=Fusarium vanettenii OX=2747968 GN=CUT2 PE=3 SV=1 |
|
| 2.05e-69 | 1 | 244 | 1 | 226 | Cutinase 3 OS=Fusarium vanettenii OX=2747968 GN=CUT3 PE=3 SV=1 |
|
| 5.80e-69 | 50 | 245 | 14 | 209 | Cutinase OS=Alternaria brassicicola OX=29001 GN=CUTAB1 PE=2 SV=1 |
|
| 9.19e-68 | 1 | 244 | 1 | 225 | Cutinase 1 OS=Fusarium vanettenii OX=2747968 GN=CUT1 PE=1 SV=1 |
|
| 5.25e-67 | 1 | 244 | 1 | 225 | Cutinase OS=Fusarium solani subsp. cucurbitae OX=2747967 GN=CUTA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
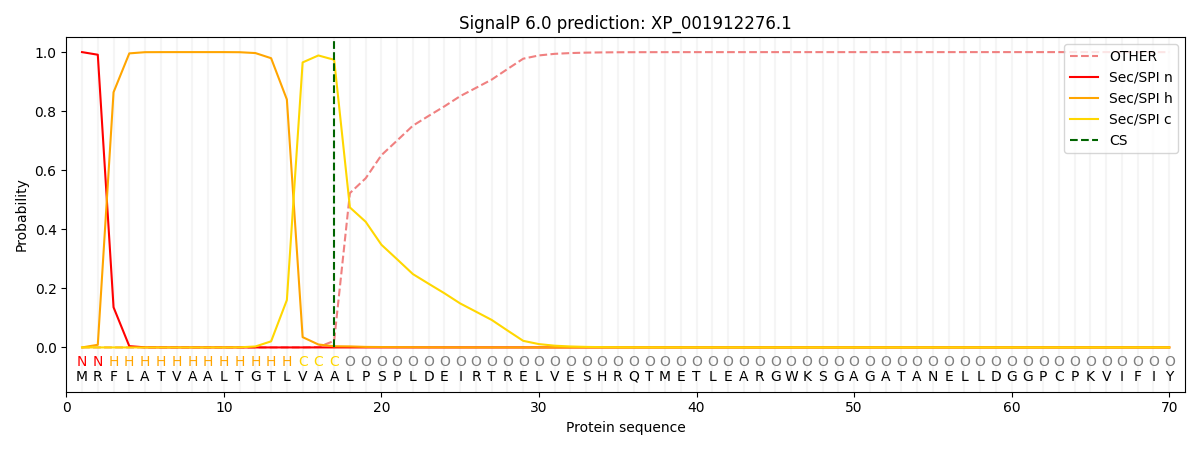
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000405 | 0.999558 | CS pos: 17-18. Pr: 0.9740 |
