You are browsing environment: FUNGIDB
CAZyme Information: XP_001911494.1
You are here: Home > Sequence: XP_001911494.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001911494.1 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | Cellulase domain-containing protein [Source:UniProtKB/TrEMBL;Acc:B2B694] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 22 | 320 | 9.6e-86 | 0.9858156028368794 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 3.90e-36 | 111 | 318 | 81 | 271 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 2.28e-07 | 111 | 323 | 132 | 367 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.87e-292 | 1 | 382 | 1 | 382 | |
| 2.87e-292 | 1 | 382 | 1 | 382 | |
| 1.76e-260 | 1 | 382 | 1 | 387 | |
| 5.11e-192 | 9 | 381 | 12 | 389 | |
| 1.01e-189 | 9 | 381 | 12 | 390 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.26e-50 | 13 | 337 | 15 | 323 | Structure of XacCel5A crystallized in the space group P41212 [Xanthomonas citri pv. citri str. 306] |
|
| 1.55e-50 | 13 | 337 | 15 | 323 | Crystal structure of XacCel5A in the native form [Xanthomonas citri pv. citri str. 306],4W7V_A Crystal structure of XacCel5A in complex with cellobiose [Xanthomonas citri pv. citri str. 306],4W7W_A High-resolution structure of XacCel5A in complex with cellopentaose [Xanthomonas citri pv. citri str. 306] |
|
| 2.82e-48 | 13 | 337 | 15 | 323 | Crystal structure of the endo-beta-1,4-glucanase (Xac0030) from Xanthomonas axonopodis pv. citri with the triple mutation His174Trp, Tyr211Ala and Lys227Arg. [Xanthomonas citri pv. citri str. 306],5HNN_B Crystal structure of the endo-beta-1,4-glucanase (Xac0030) from Xanthomonas axonopodis pv. citri with the triple mutation His174Trp, Tyr211Ala and Lys227Arg. [Xanthomonas citri pv. citri str. 306],5HNN_C Crystal structure of the endo-beta-1,4-glucanase (Xac0030) from Xanthomonas axonopodis pv. citri with the triple mutation His174Trp, Tyr211Ala and Lys227Arg. [Xanthomonas citri pv. citri str. 306] |
|
| 4.78e-47 | 32 | 320 | 5 | 277 | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger [Aspergillus niger] |
|
| 3.83e-46 | 32 | 320 | 7 | 279 | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger],5I79_B Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.76e-169 | 13 | 381 | 12 | 384 | Endoglucanase 1 OS=Robillarda sp. (strain Y-20) OX=72589 GN=eg 1 PE=1 SV=2 |
|
| 2.79e-47 | 19 | 337 | 114 | 421 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
|
| 1.36e-46 | 1 | 320 | 1 | 307 | Probable endo-beta-1,4-glucanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=eglB PE=3 SV=1 |
|
| 3.66e-46 | 1 | 320 | 1 | 306 | Endo-beta-1,4-glucanase B OS=Aspergillus niger OX=5061 GN=eglB PE=1 SV=1 |
|
| 3.66e-46 | 1 | 320 | 1 | 306 | Probable endo-beta-1,4-glucanase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
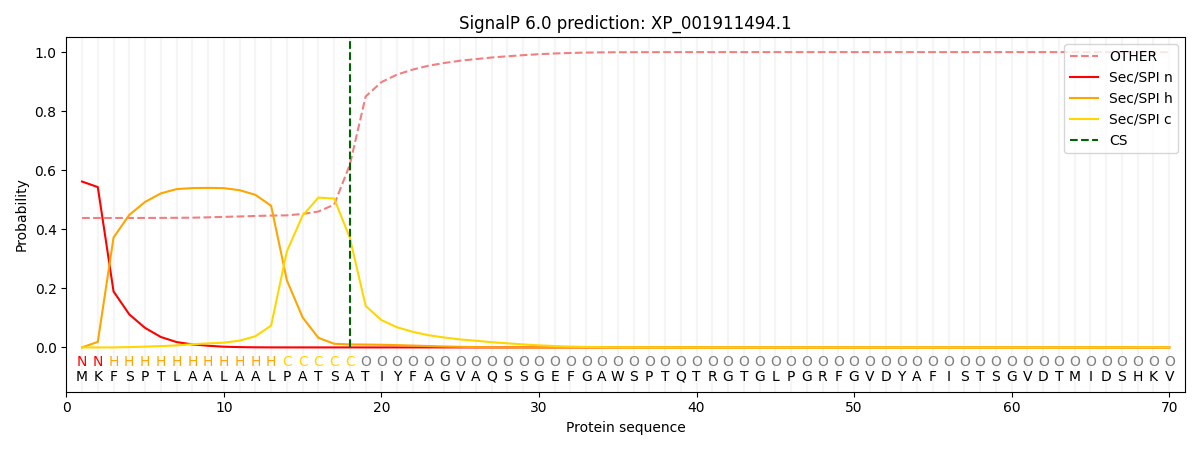
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.447388 | 0.552614 | CS pos: 18-19. Pr: 0.3703 |
