You are browsing environment: FUNGIDB
CAZyme Information: XP_001911063.1
Basic Information
help
| Species |
Podospora anserina
|
| Lineage |
Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina
|
| CAZyme ID |
XP_001911063.1
|
| CAZy Family |
GH72 |
| CAZyme Description |
Podospora anserina S mat+ genomic DNA chromosome 2, supercontig 2 [Source:UniProtKB/TrEMBL;Acc:B2B513]
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 791 |
|
87365.51 |
5.1407 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PanserinaSmat |
10888 |
515849 |
370 |
10518
|
|
| Gene Location |
No EC number prediction in XP_001911063.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH78 |
269 |
689 |
5.3e-46 |
0.7242063492063492 |
XP_001911063.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
791 |
1 |
791 |
| 0.0 |
1 |
791 |
1 |
791 |
| 0.0 |
1 |
791 |
1 |
793 |
| 6.04e-244 |
21 |
772 |
42 |
809 |
| 7.80e-156 |
21 |
690 |
31 |
648 |
XP_001911063.1 has no PDB hit.
XP_001911063.1 has no Swissprot hit.
This protein is predicted as SP
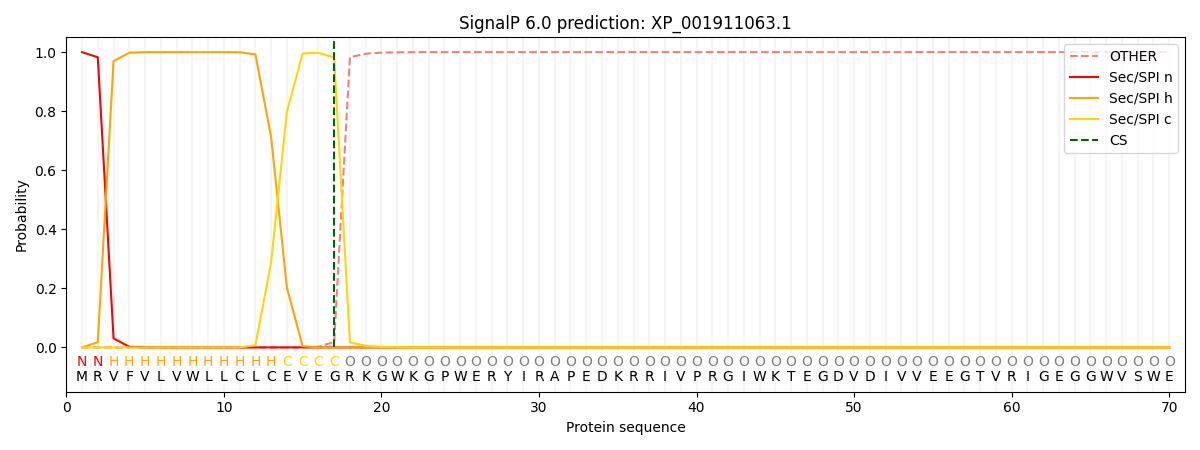
| Other |
SP_Sec_SPI |
CS Position |
| 0.000269 |
0.999699 |
CS pos: 17-18. Pr: 0.9817 |
There is no transmembrane helices in XP_001911063.1.

