You are browsing environment: FUNGIDB
CAZyme Information: XP_001910837.1
You are here: Home > Sequence: XP_001910837.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001910837.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | Podospora anserina S mat+ genomic DNA chromosome 2, supercontig 2 [Source:UniProtKB/TrEMBL;Acc:B2B4D7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:12 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 3.58e-36 | 651 | 854 | 1 | 199 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 223021 | PHA03247 | 0.001 | 425 | 640 | 2710 | 2918 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 864 | 1 | 864 | |
| 0.0 | 1 | 864 | 1 | 864 | |
| 0.0 | 1 | 864 | 1 | 963 | |
| 8.18e-30 | 649 | 864 | 28 | 249 | |
| 3.27e-27 | 649 | 864 | 22 | 239 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.05e-23 | 649 | 859 | 4 | 210 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 7.93e-18 | 651 | 842 | 1 | 204 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 8.05e-18 | 651 | 842 | 2 | 205 | ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XTK_B ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XUC_A Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_B Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_C Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XVP_A ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163],2XVP_B ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163] |
|
| 8.05e-18 | 651 | 842 | 2 | 205 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
|
| 4.80e-14 | 653 | 864 | 3 | 202 | Chain A, Hevamine A [Hevea brasiliensis],1KQZ_A Chain A, Hevamine A [Hevea brasiliensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.49e-23 | 646 | 842 | 18 | 215 | Chitinase 3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CHT3 PE=1 SV=2 |
|
| 5.31e-22 | 645 | 859 | 21 | 231 | Endochitinase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CTS1 PE=1 SV=2 |
|
| 3.47e-20 | 645 | 864 | 22 | 256 | Endochitinase A OS=Emericella nidulans OX=162425 GN=chiA PE=2 SV=3 |
|
| 3.47e-20 | 645 | 864 | 22 | 256 | Endochitinase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=chiA PE=1 SV=1 |
|
| 3.57e-20 | 649 | 864 | 27 | 246 | Endochitinase 3 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
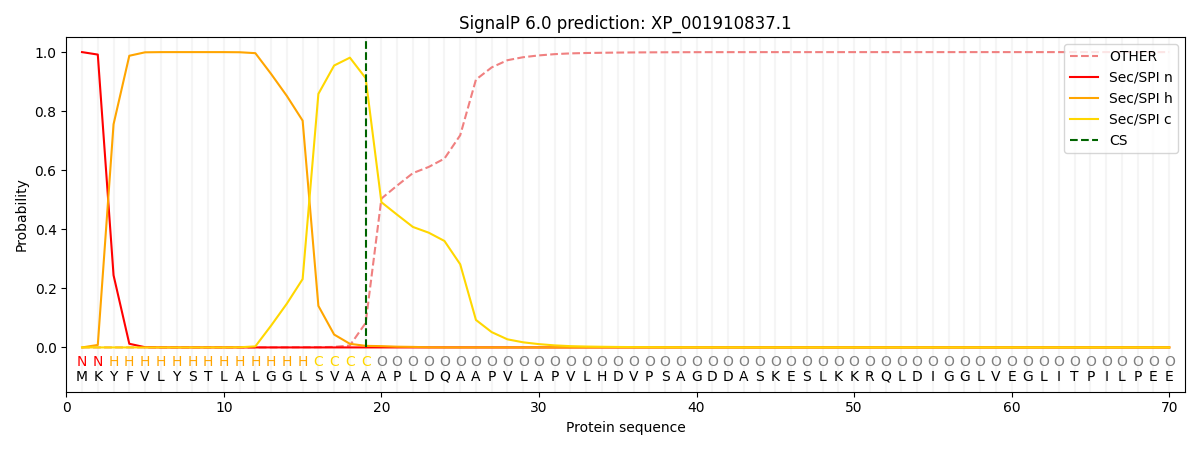
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000331 | 0.999633 | CS pos: 19-20. Pr: 0.9118 |
