You are browsing environment: FUNGIDB
CAZyme Information: XP_001910739.1
You are here: Home > Sequence: XP_001910739.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001910739.1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | FAD-oxidoreductase [Source:UniProtKB/TrEMBL;Acc:B2B439] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.3.10:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 16 | 598 | 4e-248 | 0.9872495446265938 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274143 | pyranose_ox | 0.0 | 17 | 600 | 5 | 547 | pyranose oxidase. Pyranose oxidase (also called glucose 2-oxidase) converts D-glucose and molecular oxygen to 2-dehydro-D-glucose and hydrogen peroxide. Peroxide production is believed to be important to the wood rot fungi in which this enzyme is found for lignin degradation. |
| 398739 | GMC_oxred_C | 1.58e-14 | 468 | 591 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 225186 | BetA | 1.89e-13 | 8 | 595 | 7 | 533 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 366272 | GMC_oxred_N | 5.41e-04 | 219 | 303 | 127 | 202 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 607 | 1 | 607 | |
| 0.0 | 1 | 607 | 1 | 607 | |
| 0.0 | 1 | 605 | 1 | 587 | |
| 1.13e-207 | 8 | 595 | 5 | 573 | |
| 2.35e-202 | 8 | 595 | 5 | 595 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.25e-123 | 17 | 595 | 51 | 609 | Chain A, Pyranose oxidase [Trametes ochracea],3BG7_B Chain B, Pyranose oxidase [Trametes ochracea],3BG7_C Chain C, Pyranose oxidase [Trametes ochracea],3BG7_D Chain D, Pyranose oxidase [Trametes ochracea],3BG7_E Chain E, Pyranose oxidase [Trametes ochracea],3BG7_F Chain F, Pyranose oxidase [Trametes ochracea],3BG7_G Chain G, Pyranose oxidase [Trametes ochracea],3BG7_H Chain H, Pyranose oxidase [Trametes ochracea] |
|
| 2.78e-123 | 17 | 595 | 23 | 581 | Reaction geometry and thermostability mutant of pyranose 2-oxidase from the white-rot fungus Peniophora sp. [Peniophora sp. SG] |
|
| 3.17e-123 | 17 | 595 | 51 | 609 | Crystal Structure of Pyranose 2-Oxidase [Trametes ochracea],1TT0_B Crystal Structure of Pyranose 2-Oxidase [Trametes ochracea],1TT0_C Crystal Structure of Pyranose 2-Oxidase [Trametes ochracea],1TT0_D Crystal Structure of Pyranose 2-Oxidase [Trametes ochracea],2IGK_A Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_B Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_C Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_D Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_E Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_F Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_G Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea],2IGK_H Crystal structure of recombinant pyranose 2-oxidase [Trametes ochracea] |
|
| 4.22e-123 | 17 | 595 | 51 | 609 | Pyranose 2-oxidase V546C mutant with 3-fluorinated glucose [Trametes ochracea],4MOH_A Pyranose 2-oxidase V546C mutant with 2-fluorinated glucose [Trametes ochracea],4MOP_A Pyranose 2-oxidase V546C mutant with 3-fluorinated galactose [Trametes ochracea],4MOP_B Pyranose 2-oxidase V546C mutant with 3-fluorinated galactose [Trametes ochracea],4MOP_C Pyranose 2-oxidase V546C mutant with 3-fluorinated galactose [Trametes ochracea],4MOP_D Pyranose 2-oxidase V546C mutant with 3-fluorinated galactose [Trametes ochracea],4MOQ_A Pyranose 2-oxidase V546C mutant with 2-fluorinated galactose [Trametes ochracea],4MOQ_B Pyranose 2-oxidase V546C mutant with 2-fluorinated galactose [Trametes ochracea] |
|
| 1.10e-122 | 17 | 595 | 23 | 581 | Chain A, Pyranose 2-oxidase [Peniophora sp. SG] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.39e-164 | 17 | 600 | 27 | 579 | Pyranose 2-oxidase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=p2ox PE=3 SV=2 |
|
| 3.23e-122 | 17 | 595 | 51 | 609 | Pyranose 2-oxidase OS=Trametes versicolor OX=5325 GN=P2OX PE=1 SV=1 |
|
| 3.23e-122 | 17 | 595 | 51 | 609 | Pyranose 2-oxidase OS=Peniophora sp. (strain SG) OX=204723 GN=p2ox PE=1 SV=1 |
|
| 6.13e-121 | 17 | 604 | 40 | 610 | Pyranose 2-oxidase OS=Lyophyllum shimeji OX=47721 GN=p2ox PE=2 SV=1 |
|
| 2.11e-119 | 17 | 595 | 51 | 609 | Pyranose 2-oxidase OS=Trametes pubescens OX=154538 GN=p2ox PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
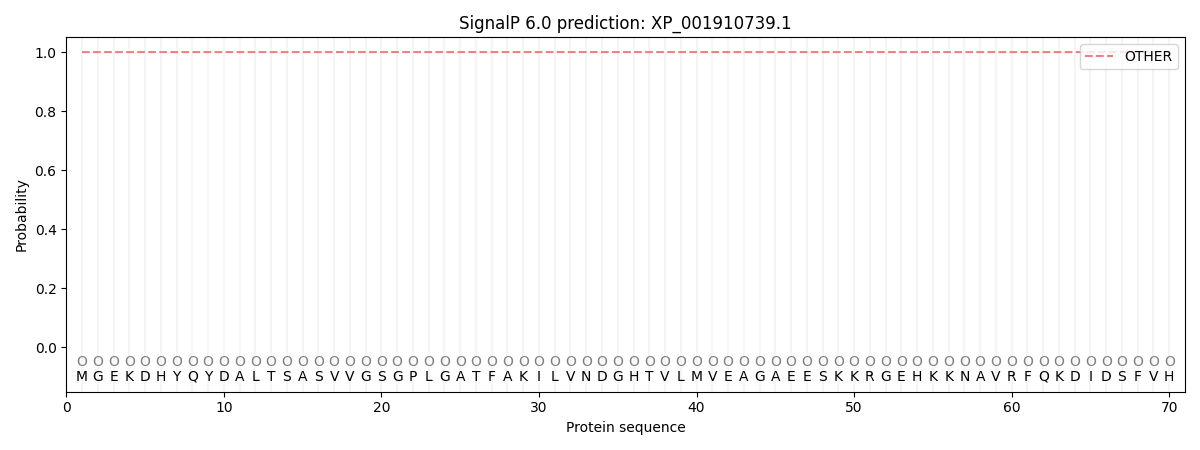
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000059 | 0.000000 |
