You are browsing environment: FUNGIDB
CAZyme Information: XP_001909162.1
You are here: Home > Sequence: XP_001909162.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001909162.1 | |||||||||||
| CAZy Family | GH43|CBM6 | |||||||||||
| CAZyme Description | Glucoamylase [Source:UniProtKB/TrEMBL;Acc:B2AZ14] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.3:94 | 3.2.1.3:71 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH15 | 47 | 455 | 1.6e-69 | 0.9695290858725761 |
| CBM20 | 507 | 597 | 4.7e-22 | 0.9333333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395586 | Glyco_hydro_15 | 9.54e-124 | 38 | 455 | 1 | 413 | Glycosyl hydrolases family 15. In higher organisms this family is represented by phosphorylase kinase subunits. |
| 99886 | CBM20_glucoamylase | 8.65e-23 | 508 | 581 | 9 | 79 | Glucoamylase (glucan1,4-alpha-glucosidase), C-terminal CBM20 (carbohydrate-binding module, family 20) domain. Glucoamylases are inverting, exo-acting starch hydrolases that hydrolyze starch and related polysaccharides by releasing the nonreducing end glucose. They are mainly active on alpha-1,4-glycosidic bonds but also have some activity towards 1,6-glycosidic bonds occurring in natural oligosaccharides. The ability of glucoamylases to cleave 1-6-glycosidic binds is called "debranching activity" and is of importance in industrial applications, where complete degradation of starch to glucose is needed. Most glucoamylases are multidomain proteins containing an N-terminal catalytic domain, a C-terminal CBM20 domain, and a highly O-glycosylated linker region that connects the two. The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 119437 | CBM20 | 3.23e-18 | 508 | 583 | 2 | 74 | The family 20 carbohydrate-binding module (CBM20), also known as the starch-binding domain, is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 395557 | CBM_20 | 3.43e-15 | 508 | 575 | 3 | 66 | Starch binding domain. |
| 215006 | CBM_2 | 1.86e-13 | 506 | 575 | 1 | 67 | Starch binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 623 | 1 | 623 | |
| 0.0 | 1 | 623 | 1 | 623 | |
| 0.0 | 1 | 623 | 1 | 623 | |
| 1.49e-232 | 14 | 623 | 10 | 611 | |
| 1.44e-231 | 29 | 623 | 31 | 616 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.98e-233 | 29 | 623 | 31 | 616 | Chain A, Glucoamylase P [Amorphotheca resinae],6FHW_B Chain B, Glucoamylase P [Amorphotheca resinae] |
|
| 3.95e-183 | 29 | 580 | 8 | 564 | Crystal structure of Penicillium oxalicum Glucoamylase [Penicillium oxalicum 114-2] |
|
| 2.36e-171 | 29 | 583 | 3 | 590 | Structure of the catalytic domain of Aspergillus niger Glucoamylase [Aspergillus niger] |
|
| 1.54e-170 | 29 | 583 | 2 | 572 | Chain A, GLUCOAMYLASE [Trichoderma reesei],2VN7_A Chain A, GLUCOAMYLASE [Trichoderma reesei] |
|
| 1.21e-160 | 29 | 500 | 3 | 469 | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.56e-232 | 29 | 623 | 31 | 616 | Glucoamylase P OS=Amorphotheca resinae OX=5101 GN=GAMP PE=1 SV=1 |
|
| 5.33e-171 | 29 | 583 | 30 | 586 | Glucoamylase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=glaA PE=2 SV=2 |
|
| 1.31e-170 | 5 | 583 | 5 | 614 | Glucoamylase OS=Aspergillus niger OX=5061 GN=GLAA PE=1 SV=1 |
|
| 1.31e-170 | 5 | 583 | 5 | 614 | Glucoamylase OS=Aspergillus awamori OX=105351 GN=GLAA PE=1 SV=1 |
|
| 3.60e-170 | 5 | 583 | 5 | 613 | Glucoamylase OS=Aspergillus usamii OX=186680 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
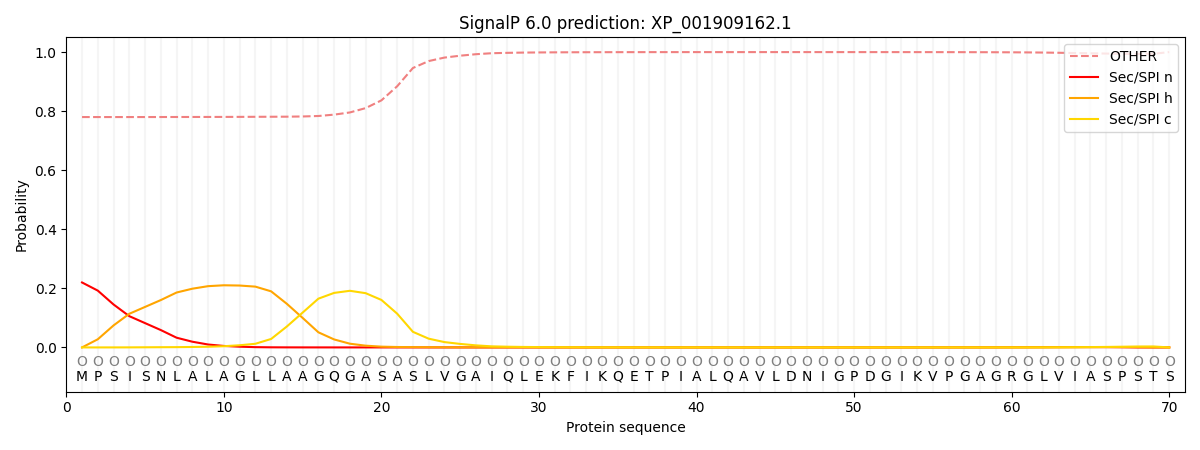
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.795598 | 0.204417 |
