You are browsing environment: FUNGIDB
CAZyme Information: XP_001907681.1
You are here: Home > Sequence: XP_001907681.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001907681.1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Podospora anserina S mat+ genomic DNA chromosome 7, supercontig 1 [Source:UniProtKB/TrEMBL;Acc:B2AVD0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 131 | 265 | 5.6e-28 | 0.8525641025641025 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 5.79e-17 | 47 | 250 | 8 | 190 | Glycosyl hydrolase family 12. |
| 235746 | PRK06215 | 1.80e-06 | 7 | 263 | 15 | 233 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.53e-237 | 1 | 320 | 1 | 320 | |
| 9.53e-237 | 1 | 320 | 1 | 320 | |
| 3.19e-235 | 1 | 320 | 1 | 320 | |
| 1.03e-92 | 25 | 291 | 1 | 270 | |
| 1.02e-67 | 23 | 276 | 20 | 250 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.39e-22 | 28 | 250 | 2 | 202 | Chain A, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride],1OA3_B Chain B, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride],1OA3_C Chain C, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride],1OA3_D Chain D, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride] |
|
| 7.01e-16 | 23 | 268 | 1 | 222 | Crystal structure of xeg-edgp [Aspergillus aculeatus],3VLB_D Crystal structure of xeg-edgp [Aspergillus aculeatus] |
|
| 1.08e-15 | 36 | 268 | 20 | 229 | Crystal structure of XEG [Aspergillus aculeatus],3VL9_A Crystal structure of xeg-xyloglucan [Aspergillus aculeatus],3VL9_B Crystal structure of xeg-xyloglucan [Aspergillus aculeatus] |
|
| 1.20e-15 | 20 | 250 | 10 | 218 | CRYSTAL STRUCTURE 4Ac Endoglucanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550],5M2D_B CRYSTAL STRUCTURE 4Ac Endoglucanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550] |
|
| 1.84e-15 | 28 | 250 | 2 | 206 | The structure of Aspergillus niger endoglucanase-palladium complex [Aspergillus niger],1KS5_A Structure of Aspergillus niger endoglucanase [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.24e-21 | 28 | 265 | 41 | 254 | Endoglucanase cel12C OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12C PE=3 SV=1 |
|
| 8.64e-19 | 23 | 263 | 33 | 252 | Endoglucanase cel12A OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12A PE=1 SV=1 |
|
| 4.08e-17 | 10 | 250 | 4 | 222 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
|
| 1.10e-16 | 19 | 250 | 14 | 224 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger OX=5061 GN=xgeA PE=1 SV=1 |
|
| 2.08e-16 | 19 | 250 | 14 | 224 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xgeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
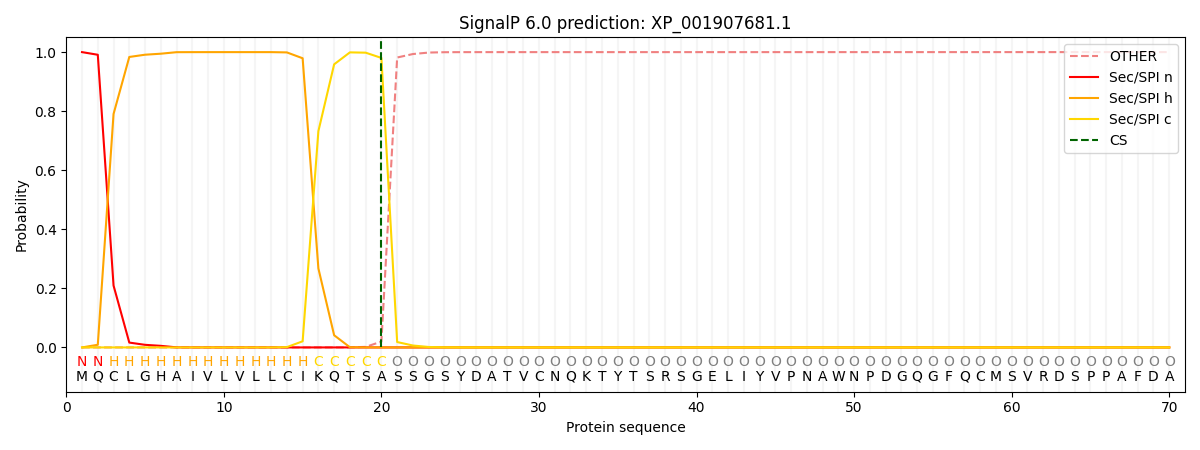
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000193 | 0.999754 | CS pos: 20-21. Pr: 0.9803 |
