You are browsing environment: FUNGIDB
CAZyme Information: XP_001907527.1
You are here: Home > Sequence: XP_001907527.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001907527.1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | GMC_OxRdtase_N domain-containing protein [Source:UniProtKB/TrEMBL;Acc:B2AUX5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 56 | 644 | 1.8e-77 | 0.5129449838187702 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 1.34e-55 | 75 | 641 | 23 | 536 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 2.75e-54 | 81 | 639 | 28 | 532 | choline dehydrogenase; Validated |
| 398739 | GMC_oxred_C | 4.48e-25 | 497 | 634 | 4 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 366272 | GMC_oxred_N | 7.24e-20 | 153 | 421 | 19 | 215 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 215420 | PLN02785 | 4.90e-14 | 313 | 630 | 234 | 568 | Protein HOTHEAD |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 658 | 1 | 658 | |
| 0.0 | 1 | 658 | 1 | 658 | |
| 8.11e-174 | 39 | 631 | 13 | 590 | |
| 2.77e-161 | 36 | 641 | 10 | 665 | |
| 9.36e-160 | 52 | 641 | 32 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.96e-28 | 59 | 641 | 5 | 530 | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state [Methylovorus sp. MP688],4UDP_B Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the oxidized state [Methylovorus sp. MP688],4UDQ_A Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state [Methylovorus sp. MP688],4UDQ_B Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state [Methylovorus sp. MP688] |
|
| 1.96e-28 | 59 | 641 | 5 | 530 | Crystal structure of the V465T mutant of 5-(Hydroxymethyl)furfural Oxidase (HMFO) [Methylovorus sp. MP688],6F97_B Crystal structure of the V465T mutant of 5-(Hydroxymethyl)furfural Oxidase (HMFO) [Methylovorus sp. MP688] |
|
| 2.02e-27 | 59 | 641 | 5 | 530 | Crystal structure of the H467A mutant of 5-hydroxymethylfurfural oxidase (HMFO) [Methylovorus sp. MP688],4UDR_B Crystal structure of the H467A mutant of 5-hydroxymethylfurfural oxidase (HMFO) [Methylovorus sp. MP688] |
|
| 1.00e-26 | 56 | 635 | 37 | 594 | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype [Leucoagaricus meleagris] |
|
| 1.72e-26 | 60 | 636 | 14 | 524 | Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_B Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_C Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_D Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_E Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_F Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_G Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_H Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.43e-40 | 60 | 636 | 3 | 529 | Oxygen-dependent choline dehydrogenase OS=Yersinia pestis (strain Pestoides F) OX=386656 GN=betA PE=3 SV=1 |
|
| 5.06e-40 | 48 | 636 | 34 | 574 | Choline dehydrogenase, mitochondrial OS=Rattus norvegicus OX=10116 GN=Chdh PE=1 SV=1 |
|
| 6.34e-40 | 60 | 636 | 3 | 529 | Oxygen-dependent choline dehydrogenase OS=Yersinia pseudotuberculosis serotype O:3 (strain YPIII) OX=502800 GN=betA PE=3 SV=1 |
|
| 6.34e-40 | 60 | 636 | 3 | 529 | Oxygen-dependent choline dehydrogenase OS=Yersinia pestis bv. Antiqua (strain Nepal516) OX=377628 GN=betA PE=3 SV=1 |
|
| 6.34e-40 | 60 | 636 | 3 | 529 | Oxygen-dependent choline dehydrogenase OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=betA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
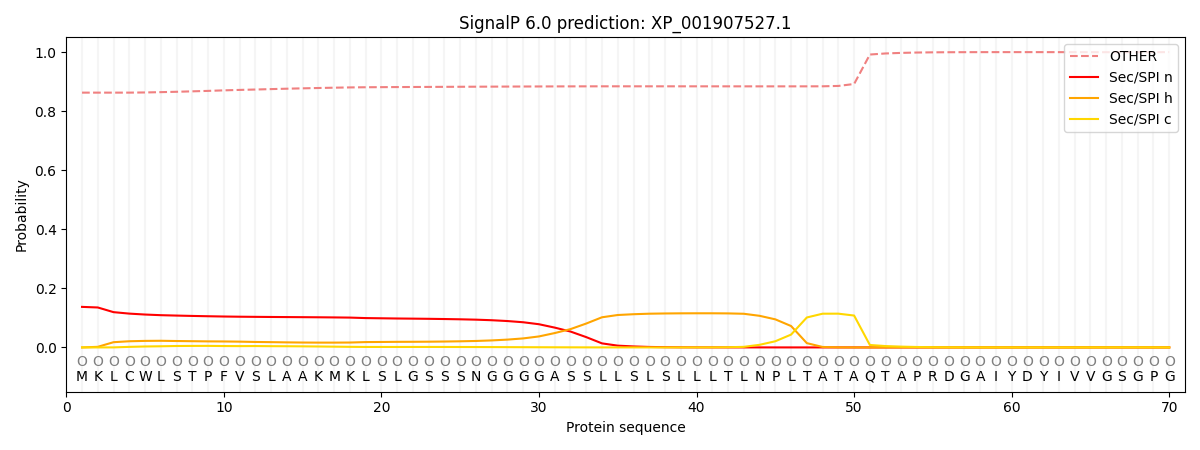
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.866366 | 0.133634 |
