You are browsing environment: FUNGIDB
CAZyme Information: XP_001906957.1
You are here: Home > Sequence: XP_001906957.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001906957.1 | |||||||||||
| CAZy Family | GH15 | |||||||||||
| CAZyme Description | CBM1 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:B2ATA5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 21 | 212 | 1e-54 | 0.9639175257731959 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 1.72e-49 | 21 | 211 | 1 | 150 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 404371 | Lipase_GDSL_2 | 2.80e-16 | 25 | 206 | 1 | 173 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238141 | SGNH_hydrolase | 5.27e-13 | 23 | 217 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 197593 | fCBD | 4.76e-11 | 266 | 298 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 238866 | sialate_O-acetylesterase_like2 | 1.06e-09 | 98 | 206 | 46 | 156 | sialate_O-acetylesterase_like subfamily of the SGNH-hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.01e-220 | 1 | 298 | 1 | 298 | |
| 1.01e-220 | 1 | 298 | 1 | 298 | |
| 4.12e-220 | 1 | 298 | 1 | 298 | |
| 6.39e-164 | 8 | 298 | 6 | 286 | |
| 5.20e-132 | 1 | 298 | 1 | 283 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.59e-128 | 21 | 225 | 2 | 206 | Crystal structure of a carbohydrate esterase family 3 from Talaromyces cellulolyticus [Talaromyces cellulolyticus] |
|
| 2.46e-127 | 21 | 225 | 2 | 206 | Crystal structure of acetyl esterase mutant S10A with acetate ion [Talaromyces cellulolyticus] |
|
| 1.36e-17 | 17 | 211 | 2 | 188 | Chain A, LIPOLYTIC ENZYME [Acetivibrio thermocellus] |
|
| 2.05e-06 | 265 | 298 | 2 | 36 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 5.28e-06 | 268 | 298 | 5 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.18e-15 | 262 | 298 | 259 | 295 | Cutinase 11 OS=Verticillium dahliae OX=27337 GN=VD0003_g7577 PE=1 SV=1 |
|
| 3.49e-15 | 261 | 298 | 273 | 310 | Endo-1,4-beta-xylanase B OS=Penicillium oxalicum OX=69781 GN=xynB PE=1 SV=1 |
|
| 1.97e-13 | 261 | 298 | 392 | 429 | Probable exoglucanase GH6D OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=GH6D PE=3 SV=1 |
|
| 2.99e-13 | 255 | 298 | 248 | 290 | Endo-1,4-beta-xylanase B OS=Phanerodontia chrysosporium OX=2822231 GN=xynB PE=1 SV=1 |
|
| 1.64e-12 | 263 | 297 | 274 | 308 | Endoglucanase cel12C OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12C PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
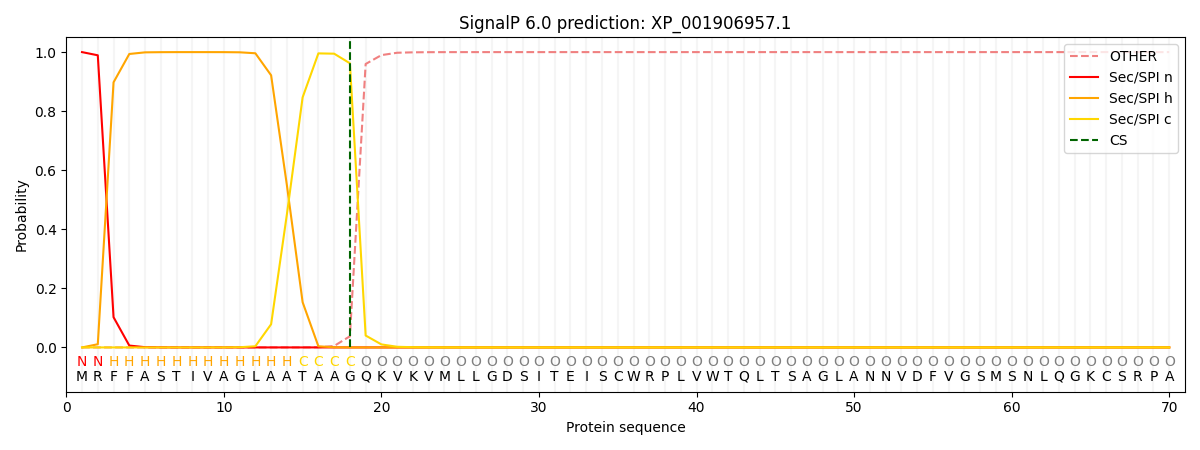
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000481 | 0.999496 | CS pos: 18-19. Pr: 0.9621 |
