You are browsing environment: FUNGIDB
CAZyme Information: XP_001906104.1
You are here: Home > Sequence: XP_001906104.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
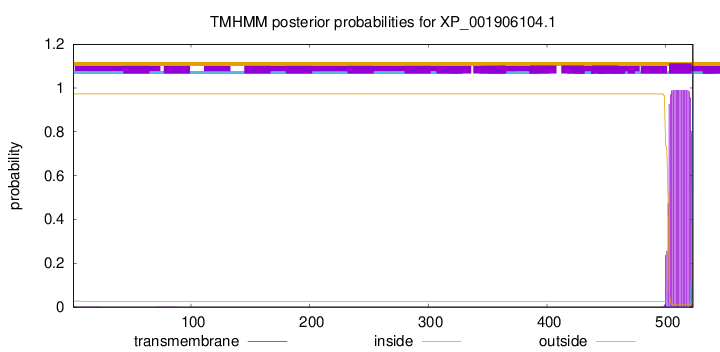
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001906104.1 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | Podospora anserina S mat+ genomic DNA chromosome 4, supercontig 4 [Source:UniProtKB/TrEMBL;Acc:B2ARJ2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 40 | 438 | 1.9e-160 | 0.9825436408977556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 7.15e-15 | 8 | 436 | 14 | 392 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| 400653 | SGL | 0.004 | 103 | 153 | 125 | 177 | SMP-30/Gluconolaconase/LRE-like region. This family describes a region that is found in proteins expressed by a variety of eukaryotic and prokaryotic species. These proteins include various enzymes, such as senescence marker protein 30 (SMP-30), gluconolactonase and luciferin-regenerating enzyme (LRE). SMP-30 is known to hydrolyze diisopropyl phosphorofluoridate in the liver, and has been noted as having sequence similarity, in the region described in this family, with PON1 and LRE. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 523 | 1 | 523 | |
| 0.0 | 1 | 523 | 1 | 523 | |
| 0.0 | 1 | 523 | 1 | 516 | |
| 5.44e-143 | 30 | 518 | 32 | 508 | |
| 1.41e-139 | 35 | 443 | 38 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.06e-119 | 38 | 441 | 3 | 403 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 7.30e-119 | 38 | 441 | 4 | 404 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 7.54e-119 | 38 | 441 | 5 | 405 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 4.01e-69 | 41 | 440 | 11 | 410 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.38e-08 | 223 | 436 | 193 | 372 | Uncharacterized protein BB_0024 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0024 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000395 | 0.999579 | CS pos: 18-19. Pr: 0.9759 |