You are browsing environment: FUNGIDB
CAZyme Information: XP_001904121.1
You are here: Home > Sequence: XP_001904121.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001904121.1 | |||||||||||
| CAZy Family | GT66 | |||||||||||
| CAZyme Description | Podospora anserina S mat+ genomic DNA chromosome 5, supercontig 1 [Source:UniProtKB/TrEMBL;Acc:B2AEN2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 380801 | PFM_jacalin-like | 8.08e-13 | 192 | 338 | 1 | 150 | pore-forming module of uncharacterized proteins which have an N-terminal jacalin-like lectin domain, and similar aerolysin-type beta-barrel pore-forming proteins. Jacalin-like lectins are sugar-binding protein domains. Proteins having these lectin domains may bind mono- or oligosaccharides with high specificity. Generally, pore-forming proteins (PFPs) are secreted as water-soluble monomers, which upon binding to target lipid membranes, oligomerize and form transmembrane pores detrimental to cells. Beta-PFPs form pores by transmembrane beta-barrels. Aerolysin-type beta-PFPs are believed to use an amphipathic beta-hairpin to form the beta-barrel. Many of this family are bacterial toxins. A significant portion of the monomeric subunit structure is re-organized to form the pore. Oligomers formed by members of the aerolysin family include: hepta- (aerolysin), octa- (Dln1), and nonameric oligomers (lysenin and monalysin). |
| 187706 | Jacalin_like | 3.01e-08 | 40 | 163 | 1 | 121 | Jacalin-like lectin domain. Jacalin-like lectins are sugar-binding protein domains mostly found in plants. They adopt a beta-prism topology consistent with a circularly permuted three-fold repeat of a structural motif. Proteins containing this domain may bind mono- or oligosaccharides with high specificity. The domain can occur in tandem-repeat arrangements with up to six copies, and in architectures combined with a variety of other functional domains. Taxonomic distribution is not restricted to plants, the domain is also found in various mammalian proteins, for example. |
| 187711 | Jacalin_EEP | 4.21e-06 | 36 | 172 | 1 | 134 | Jacalin-like lectin domains of putative endonucleases/exonucleases/phosphatases and related proteins. Members of this taxonomically diverse family co-occur with metal-dependent endonucleases/exonucleases/phosphatases. They have not been functionally characterized. |
| 212030 | LysM | 5.26e-06 | 396 | 429 | 2 | 35 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| 187710 | griffithsin_like | 1.11e-05 | 72 | 171 | 28 | 128 | Jacalin-like lectin domain of griffithsin and related proteins. Griffithsin is a lectin isolated from a red alga, which has shown potential as an inhibitor of viral entry, exhibiting antiviral activity against HIV and SARS. The biological functions of griffithsin and griffithsin-like proteins with respect to their source organisms are not known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.32e-299 | 1 | 447 | 1 | 447 | |
| 6.75e-13 | 340 | 444 | 529 | 632 | |
| 1.54e-10 | 335 | 444 | 579 | 687 | |
| 5.98e-10 | 367 | 447 | 553 | 631 | |
| 6.05e-10 | 367 | 447 | 565 | 643 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.47e-28 | 35 | 275 | 11 | 249 | Chain A, Natterin-like protein [Lethenteron camtschaticum],5ZU4_B Chain B, Natterin-like protein [Lethenteron camtschaticum],6IUL_A Chain A, Natterin-like protein [Lethenteron camtschaticum],6IUL_B Chain B, Natterin-like protein [Lethenteron camtschaticum] |
|
| 2.93e-25 | 35 | 333 | 30 | 326 | Crystal structure of Dln1 complexed with sucrose [Danio rerio],4ZNO_B Crystal structure of Dln1 complexed with sucrose [Danio rerio],4ZNQ_A Crystal structure of Dln1 complexed with Man(alpha1-2)Man [Danio rerio],4ZNQ_B Crystal structure of Dln1 complexed with Man(alpha1-2)Man [Danio rerio],4ZNR_A Crystal structure of Dln1 complexed with Man(alpha1-3)Man [Danio rerio],4ZNR_B Crystal structure of Dln1 complexed with Man(alpha1-3)Man [Danio rerio],5DI0_A Crystal structure of Dln1 [Danio rerio],5DI0_B Crystal structure of Dln1 [Danio rerio] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.08e-24 | 35 | 333 | 10 | 306 | Aerolysin-like protein OS=Danio rerio OX=7955 GN=aep1 PE=1 SV=1 |
|
| 5.57e-10 | 386 | 445 | 380 | 437 | Probable peptidoglycan-N-acetylglucosamine deacetylase ARB_03699 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_03699 PE=1 SV=2 |
|
| 1.96e-07 | 380 | 443 | 355 | 419 | LysM domain-containing protein ARB_01155/01156 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_01155/01156 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
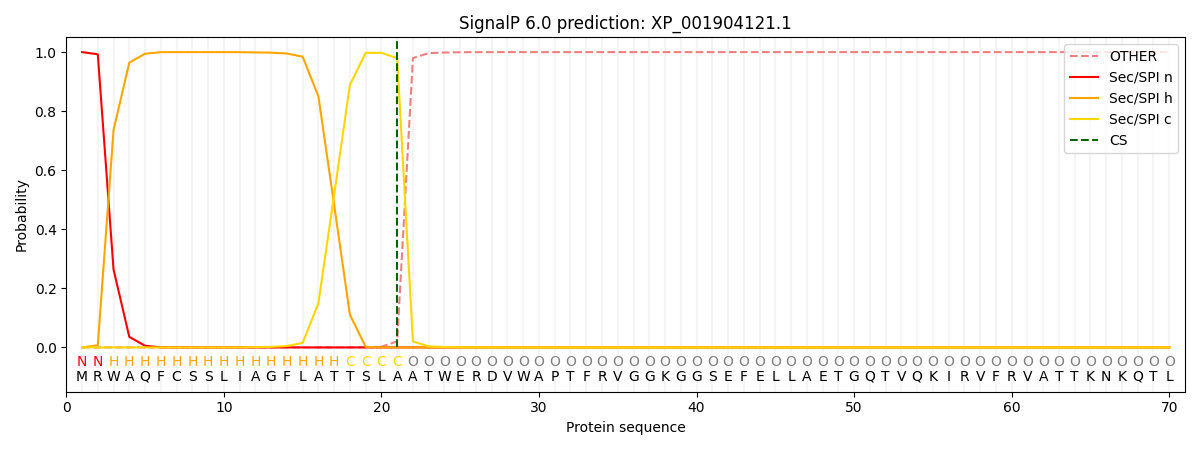
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000457 | 0.999530 | CS pos: 21-22. Pr: 0.9794 |
