You are browsing environment: FUNGIDB
CAZyme Information: XP_001903170.1
You are here: Home > Sequence: XP_001903170.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora anserina | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora anserina | |||||||||||
| CAZyme ID | XP_001903170.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | 1,4-beta-D-glucan cellobiohydrolase CEL6A [Source:UniProtKB/Swiss-Prot;Acc:B2ABX7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.91:55 | 3.2.1.4:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 153 | 445 | 1.4e-89 | 0.9319727891156463 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396075 | Glyco_hydro_6 | 7.02e-154 | 167 | 448 | 15 | 293 | Glycosyl hydrolases family 6. |
| 227616 | CelA1 | 4.36e-41 | 109 | 482 | 2 | 441 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| 197593 | fCBD | 1.50e-14 | 30 | 62 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 4.27e-14 | 31 | 58 | 2 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 484 | 1 | 484 | |
| 0.0 | 1 | 484 | 1 | 484 | |
| 0.0 | 1 | 484 | 1 | 484 | |
| 1.45e-265 | 1 | 484 | 1 | 482 | |
| 1.79e-254 | 1 | 484 | 1 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.32e-227 | 121 | 484 | 1 | 364 | Chain A, Cellobiohydrolase Family 6 [Thermochaetoides thermophila] |
|
| 1.01e-213 | 124 | 484 | 1 | 362 | Structure of the wild-type cellobiohydrolase Cel6A from Humicolas insolens in complex with a fluorescent substrate [Humicola insolens],1OCB_B Structure of the wild-type cellobiohydrolase Cel6A from Humicolas insolens in complex with a fluorescent substrate [Humicola insolens],2BVW_A CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE [Humicola insolens],2BVW_B CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE [Humicola insolens] |
|
| 2.18e-213 | 122 | 484 | 1 | 364 | D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside [Humicola insolens],1OC6_A structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution [Humicola insolens],1OC7_A D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-tetrathio-alpha-d-cellopentoside at 1.1 angstrom resolution [Humicola insolens] |
|
| 1.54e-212 | 126 | 484 | 1 | 360 | Cellobiohydrolase Ii (Cel6a) From Humicola Insolens [Humicola insolens] |
|
| 1.66e-212 | 124 | 484 | 1 | 362 | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution [Humicola insolens],1OCN_A Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 484 | 1 | 484 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=CEL6A PE=1 SV=1 |
|
| 2.15e-247 | 7 | 484 | 6 | 476 | Exoglucanase-6A OS=Humicola insolens OX=34413 GN=cel6A PE=1 SV=1 |
|
| 4.95e-218 | 1 | 483 | 1 | 486 | 1,4-beta-D-glucan cellobiohydrolase CEL6A OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel6A PE=1 SV=1 |
|
| 5.52e-202 | 13 | 484 | 9 | 464 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=cbhC PE=3 SV=1 |
|
| 4.06e-201 | 5 | 484 | 5 | 470 | Exoglucanase 2 OS=Hypocrea jecorina OX=51453 GN=cbh2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
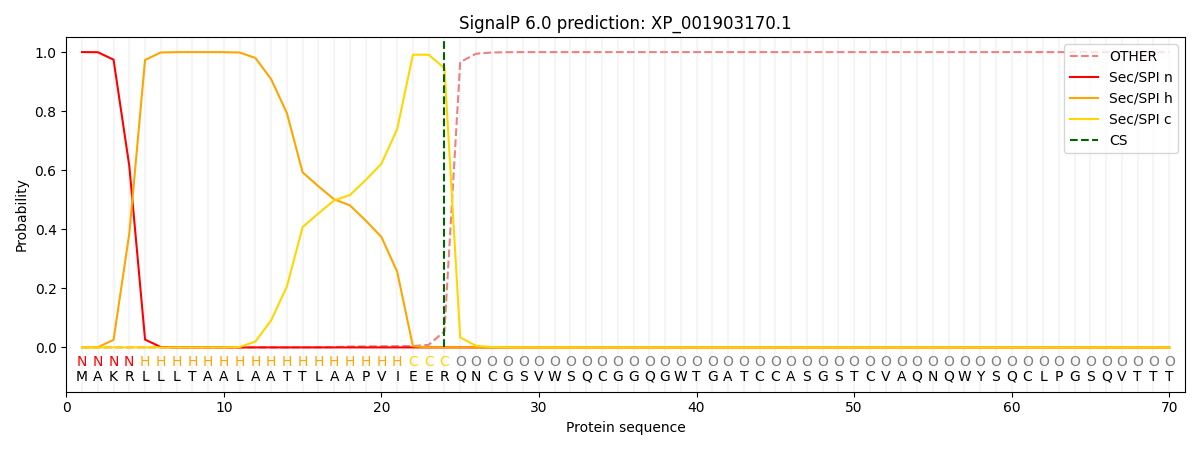
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000311 | 0.999693 | CS pos: 24-25. Pr: 0.9456 |
