You are browsing environment: FUNGIDB
CAZyme Information: VCU40439.1
You are here: Home > Sequence: VCU40439.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Blumeria graminis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Erysiphaceae; Blumeria; Blumeria graminis | |||||||||||
| CAZyme ID | VCU40439.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 43 | 374 | 2.4e-69 | 0.9022346368715084 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 9.70e-22 | 67 | 351 | 31 | 341 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 227170 | COG4833 | 1.35e-10 | 61 | 376 | 37 | 350 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.21e-287 | 1 | 379 | 1 | 379 | |
| 7.91e-236 | 1 | 379 | 1 | 379 | |
| 2.62e-197 | 10 | 379 | 10 | 379 | |
| 3.51e-88 | 40 | 378 | 26 | 379 | |
| 3.51e-88 | 40 | 378 | 26 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.67e-13 | 54 | 368 | 25 | 335 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
|
| 2.99e-13 | 72 | 368 | 60 | 353 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 4.73e-13 | 72 | 368 | 39 | 332 | Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 5.38e-13 | 72 | 368 | 73 | 366 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
|
| 1.30e-12 | 72 | 368 | 60 | 353 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5AGD_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBM_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBW_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
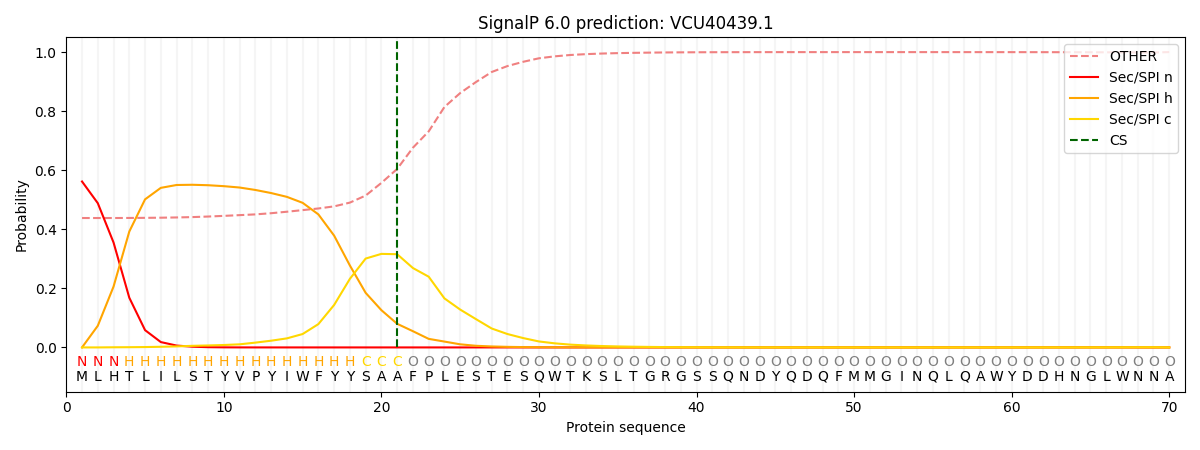
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.456128 | 0.543857 | CS pos: 21-22. Pr: 0.3156 |
