You are browsing environment: FUNGIDB
CAZyme Information: VBB80942.1
Basic Information
help
| Species |
Podospora comata
|
| Lineage |
Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora comata
|
| CAZyme ID |
VBB80942.1
|
| CAZy Family |
GH31 |
| CAZyme Description |
Putative protein of unknown function
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 263 |
LR026968|CGC12 |
28610.41 |
7.3946 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PcomataTmat |
13100 |
N/A |
2652 |
10448
|
|
| Gene Location |
No EC number prediction in VBB80942.1.
| Family |
Start |
End |
Evalue |
family coverage |
| CBM52 |
28 |
76 |
8.9e-25 |
0.9230769230769231 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 402331
|
Carb_bind |
3.94e-19 |
28 |
75 |
1 |
48 |
Carbohydrate binding. This is a carbohydrate binding domain which has been shown in Schizosaccharomyces pombe to be required for septum localization. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 7.47e-185 |
1 |
263 |
1 |
263 |
| 2.50e-183 |
1 |
263 |
1 |
263 |
| 2.50e-183 |
1 |
263 |
1 |
263 |
| 1.85e-14 |
17 |
237 |
14 |
219 |
| 1.33e-10 |
17 |
240 |
14 |
222 |
VBB80942.1 has no PDB hit.
VBB80942.1 has no Swissprot hit.
This protein is predicted as SP
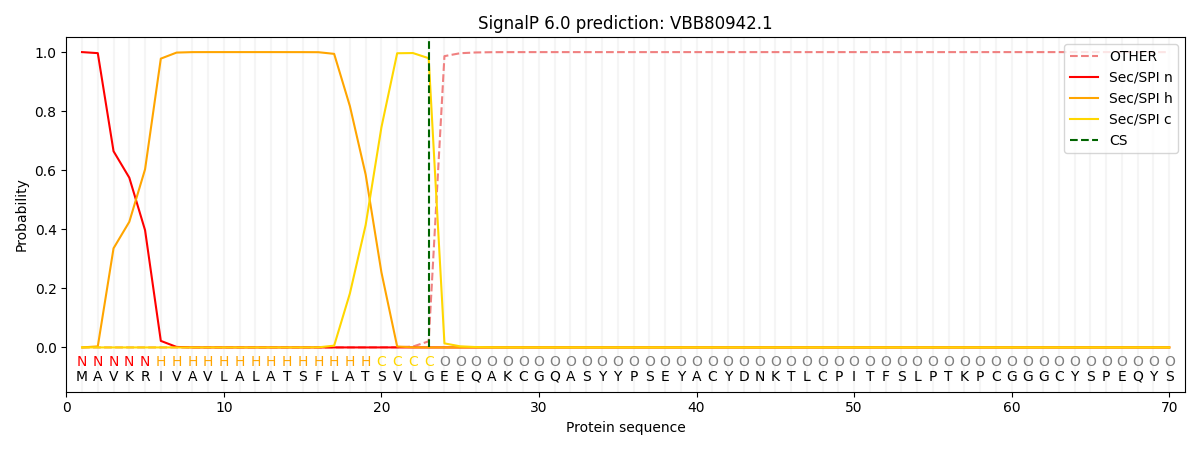
| Other |
SP_Sec_SPI |
CS Position |
| 0.000207 |
0.999775 |
CS pos: 23-24. Pr: 0.9793 |
There is no transmembrane helices in VBB80942.1.

