You are browsing environment: FUNGIDB
CAZyme Information: VBB79625.1
You are here: Home > Sequence: VBB79625.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora comata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora comata | |||||||||||
| CAZyme ID | VBB79625.1 | |||||||||||
| CAZy Family | GH131 | |||||||||||
| CAZyme Description | Putative Glycoside Hydrolase Family 10 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:45 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 62 | 356 | 1.9e-100 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 3.10e-137 | 62 | 355 | 1 | 309 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 4.03e-114 | 105 | 354 | 1 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 4.84e-73 | 35 | 359 | 1 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.28e-271 | 1 | 360 | 1 | 360 | |
| 6.50e-263 | 1 | 360 | 1 | 355 | |
| 6.50e-263 | 1 | 360 | 1 | 355 | |
| 5.22e-184 | 35 | 360 | 1 | 326 | |
| 5.99e-181 | 35 | 360 | 1 | 327 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.73e-137 | 58 | 358 | 2 | 301 | Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Trichoderma reesei [Trichoderma reesei QM6a] |
|
| 1.14e-128 | 58 | 360 | 2 | 303 | The xylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. [Thermoascus aurantiacus] |
|
| 1.62e-128 | 58 | 360 | 2 | 303 | 0.89A Ultra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
|
| 6.57e-128 | 57 | 360 | 1 | 303 | Structural studies on the mobility in the active site of the Thermoascus aurantiacus xylanase I [Thermoascus aurantiacus] |
|
| 3.77e-127 | 58 | 360 | 2 | 303 | Thermostable xylanase I from Thermoascus aurantiacus- Crystal form II [Thermoascus aurantiacus],1GOM_A Thermostable xylanase I from Thermoascus aurantiacus- Crystal form I [Thermoascus aurantiacus],1GOO_A Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex [Thermoascus aurantiacus],1GOQ_A Thermostable xylanase I from Thermoascus aurantiacus - Room temperature xylobiose complex [Thermoascus aurantiacus],1GOR_A THERMOSTABLE XYLANASE I FROM THERMOASCUS AURANTIACUS - XYLOBIOSE COMPLEX AT 100 K [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.11e-170 | 35 | 358 | 1 | 330 | Endo-1,4-beta-xylanase 2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL2 PE=3 SV=1 |
|
| 1.78e-155 | 35 | 358 | 1 | 330 | Endo-1,4-beta-xylanase 2 OS=Magnaporthe grisea OX=148305 GN=XYL2 PE=1 SV=1 |
|
| 9.70e-140 | 35 | 358 | 1 | 346 | Endo-1,4-beta-xylanase 3 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=xyn3 PE=3 SV=1 |
|
| 9.70e-140 | 35 | 358 | 1 | 346 | Endo-1,4-beta-xylanase 3 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=xyn3 PE=1 SV=1 |
|
| 1.75e-133 | 41 | 360 | 11 | 329 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
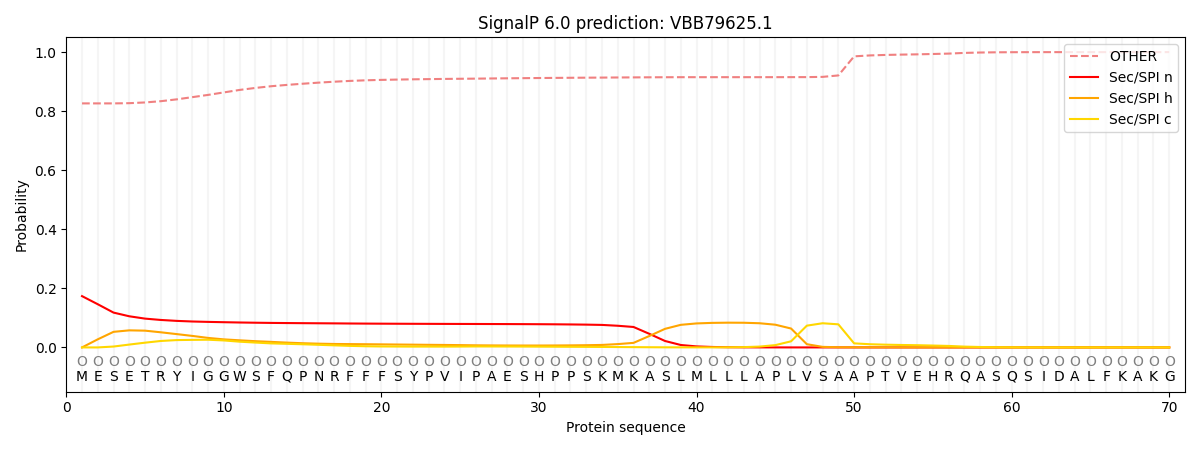
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.830915 | 0.169076 |
