You are browsing environment: FUNGIDB
CAZyme Information: VBB74964.1
You are here: Home > Sequence: VBB74964.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora comata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora comata | |||||||||||
| CAZyme ID | VBB74964.1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | Putative protein similar to Cip2 of Trichoderma reesei | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.-:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 122 | 448 | 4.2e-96 | 0.9925650557620818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395595 | CBM_1 | 5.07e-11 | 27 | 55 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 8.97e-11 | 27 | 59 | 2 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 398876 | AXE1 | 9.66e-05 | 200 | 314 | 79 | 203 | Acetyl xylan esterase (AXE1). This family consists of several bacterial acetyl xylan esterase proteins. Acetyl xylan esterases are enzymes that hydrolyze the ester linkages of the acetyl groups in position 2 and/or 3 of the xylose moieties of natural acetylated xylan from hardwood. These enzymes are one of the accessory enzymes which are part of the xylanolytic system, together with xylanases, beta-xylosidases, alpha-arabinofuranosidases and methylglucuronidases; these are all required for the complete hydrolysis of xylan. |
| 225989 | Axe1 | 0.003 | 198 | 314 | 78 | 205 | Cephalosporin-C deacetylase or related acetyl esterase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 481 | 1 | 481 | |
| 0.0 | 1 | 481 | 1 | 481 | |
| 0.0 | 1 | 481 | 1 | 481 | |
| 3.67e-180 | 5 | 480 | 4 | 495 | |
| 2.00e-163 | 22 | 480 | 16 | 459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.28e-151 | 107 | 480 | 10 | 374 | Chain A, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
|
| 2.00e-138 | 28 | 480 | 8 | 457 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 3.79e-129 | 122 | 477 | 58 | 405 | Crystal structure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
|
| 1.07e-128 | 122 | 477 | 58 | 405 | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile determined at 1.9 A resolution [Thermothelomyces thermophilus ATCC 42464],4G4J_A Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution [Thermothelomyces thermophilus ATCC 42464] |
|
| 9.11e-128 | 122 | 480 | 17 | 379 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 481 | 1 | 481 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
|
| 6.52e-181 | 5 | 480 | 4 | 495 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Sodiomyces alcalophilus OX=398408 PE=1 SV=1 |
|
| 2.48e-159 | 22 | 480 | 16 | 459 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
|
| 5.69e-139 | 117 | 480 | 37 | 393 | 4-O-methyl-glucuronoyl methylesterase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=Cip2 PE=1 SV=2 |
|
| 2.88e-138 | 21 | 480 | 17 | 473 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
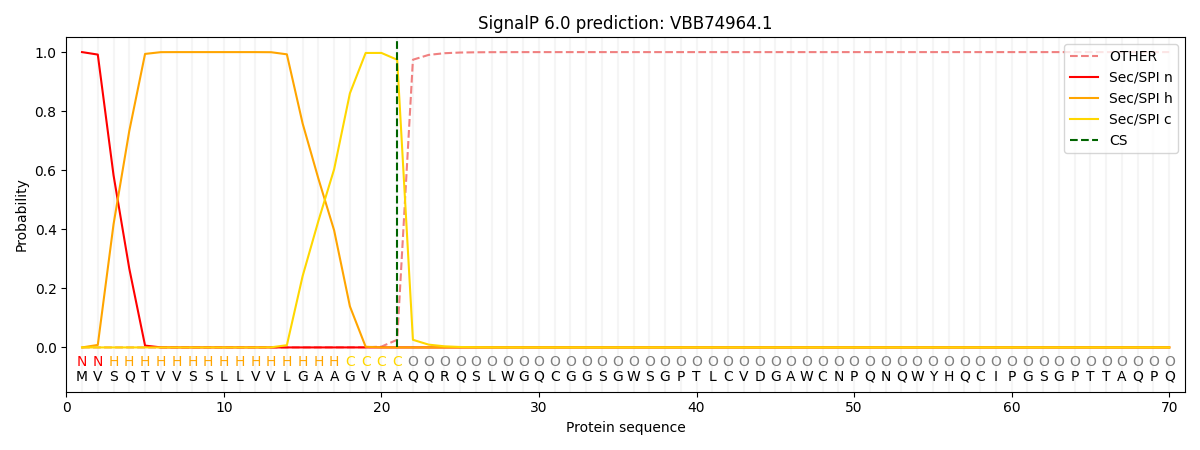
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000229 | 0.999756 | CS pos: 21-22. Pr: 0.9742 |
