You are browsing environment: FUNGIDB
CAZyme Information: VBB72572.1
You are here: Home > Sequence: VBB72572.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Podospora comata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora comata | |||||||||||
| CAZyme ID | VBB72572.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | Putative Glycoside Hydrolase Family 45 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:59 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH45 | 23 | 223 | 3.8e-90 | 0.98989898989899 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 280233 | Glyco_hydro_45 | 2.24e-80 | 22 | 223 | 1 | 213 | Glycosyl hydrolase family 45. |
| 409008 | DPBB_EXP_N-like | 0.005 | 63 | 143 | 5 | 92 | N-terminal double-psi beta-barrel fold domain of the expansin family and similar domains. The plant expansin family consists of four subfamilies, alpha-expansin (EXPA), beta-expansin (EXPB), expansin-like A (EXLA), and expansin-like B (EXLB). EXPA and EXPB display cell wall loosening activity and are involved in cell expansion and other developmental events during which cell wall modification occurs. EXPA proteins function more efficiently on dicotyledonous cell walls, whereas EXPB proteins exhibit specificity for the cell walls of monocotyledons. Expansins also affect environmental stress responses. Expansin family proteins contain an N-terminal domain (D1) homologous to the catalytic domain of glycoside hydrolase family 45 (GH45) proteins but with no hydrolytic activity, and a C-terminal domain (D2) homologous to group-2 grass pollen allergens. This family also includes GH45 endoglucanases from mollusks. This model represents the N-terminal domain of expansins and similar proteins, which adopts a double-psi beta-barrel (DPBB) fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.34e-188 | 1 | 245 | 1 | 245 | |
| 3.89e-187 | 1 | 245 | 1 | 245 | |
| 3.89e-187 | 1 | 245 | 1 | 245 | |
| 5.08e-159 | 15 | 245 | 19 | 249 | |
| 5.10e-152 | 17 | 245 | 1 | 229 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.06e-98 | 16 | 228 | 1 | 208 | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126] |
|
| 1.18e-98 | 17 | 228 | 20 | 226 | Apo Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A],6MVJ_A Cellobiose complex Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A] |
|
| 1.86e-98 | 19 | 228 | 1 | 205 | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126],5GM9_A Crystal structure of a glycoside hydrolase in complex with cellobiose [Thermothielavioides terrestris NRRL 8126] |
|
| 1.76e-96 | 23 | 228 | 3 | 204 | Endoglucanase from Humicola insolens AT 1.7A resolution [Humicola insolens] |
|
| 3.72e-95 | 23 | 228 | 3 | 204 | Endoglucanase V [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.68e-98 | 7 | 228 | 6 | 224 | Putative endoglucanase type K OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 2.12e-94 | 23 | 228 | 3 | 204 | Endoglucanase-5 OS=Humicola insolens OX=34413 PE=1 SV=1 |
|
| 5.71e-90 | 21 | 225 | 20 | 221 | Endoglucanase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
|
| 1.99e-56 | 18 | 223 | 35 | 240 | Endoglucanase OS=Phaedon cochleariae OX=80249 PE=2 SV=1 |
|
| 5.91e-43 | 23 | 221 | 269 | 496 | Endoglucanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=celB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
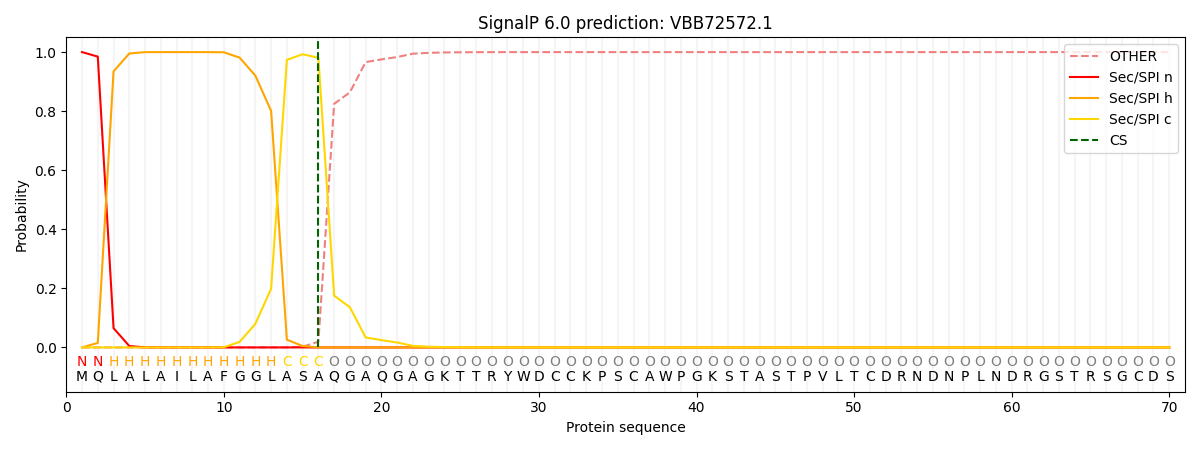
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000217 | 0.999744 | CS pos: 16-17. Pr: 0.9806 |
