You are browsing environment: FUNGIDB
CAZyme Information: VBB71571.1
Basic Information
help
| Species |
Podospora comata
|
| Lineage |
Ascomycota; Sordariomycetes; ; Podosporaceae; Podospora; Podospora comata
|
| CAZyme ID |
VBB71571.1
|
| CAZy Family |
AA12 |
| CAZyme Description |
Putative Glycosyltransferase Family 90
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 946 |
|
105879.72 |
8.9044 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PcomataTmat |
13100 |
N/A |
2652 |
10448
|
|
| Gene Location |
No EC number prediction in VBB71571.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT90 |
659 |
932 |
4.2e-49 |
0.94 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 214773
|
CAP10 |
2.10e-04 |
838 |
931 |
165 |
252 |
Putative lipopolysaccharide-modifying enzyme. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
946 |
1 |
946 |
| 0.0 |
1 |
946 |
1 |
946 |
| 0.0 |
154 |
946 |
1 |
793 |
| 6.15e-303 |
1 |
941 |
1 |
959 |
| 5.69e-256 |
153 |
941 |
2 |
797 |
VBB71571.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.16e-30 |
378 |
946 |
145 |
670 |
Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
This protein is predicted as OTHER
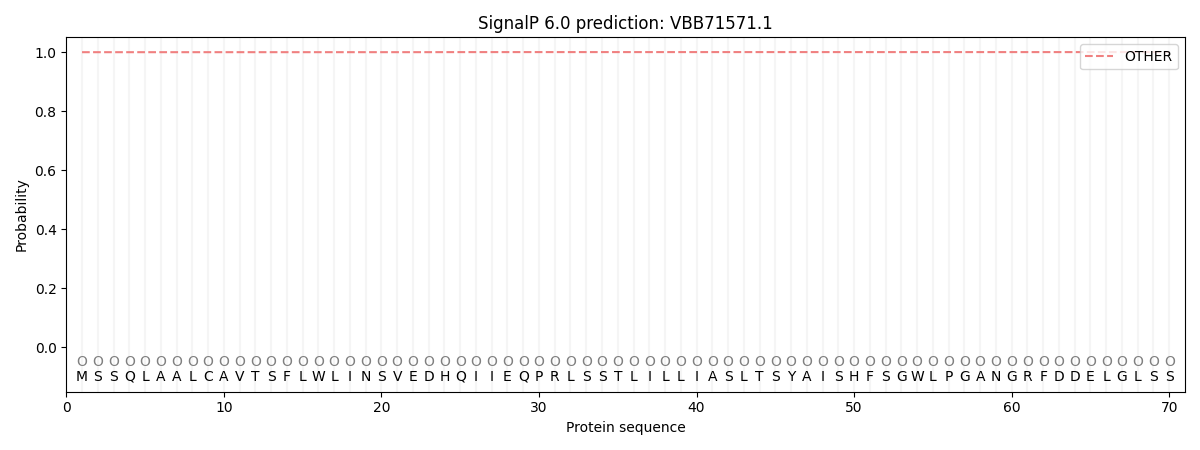
| Other |
SP_Sec_SPI |
CS Position |
| 0.999900 |
0.000128 |
|

| Start |
End |
| 32 |
54 |
| 89 |
106 |
| 163 |
185 |
| 205 |
227 |
| 239 |
256 |
| 266 |
288 |
| 295 |
317 |
| 327 |
346 |
| 353 |
375 |


