You are browsing environment: FUNGIDB
CAZyme Information: TSTA_109570-t26_1-p1
You are here: Home > Sequence: TSTA_109570-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Talaromyces stipitatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces stipitatus | |||||||||||
| CAZyme ID | TSTA_109570-t26_1-p1 | |||||||||||
| CAZy Family | GT109 | |||||||||||
| CAZyme Description | endochitinase precursor, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 32 | 296 | 9.9e-25 | 0.6993243243243243 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 1.97e-99 | 31 | 323 | 1 | 278 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 395573 | Glyco_hydro_18 | 4.69e-13 | 33 | 298 | 2 | 256 | Glycosyl hydrolases family 18. |
| 119350 | GH18_chitinase_D-like | 3.22e-07 | 33 | 291 | 3 | 268 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| 119349 | GH18_chitinase-like | 1.99e-06 | 33 | 229 | 1 | 177 | The GH18 (glycosyl hydrolase, family 18) type II chitinases hydrolyze chitin, an abundant polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) which is a major component of the cell wall of fungi and the exoskeleton of arthropods. Chitinases have been identified in viruses, bacteria, fungi, protozoan parasites, insects, and plants. The structure of the GH18 domain is an eight-stranded beta/alpha barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. The GH18 family includes chitotriosidase, chitobiase, hevamine, zymocin-alpha, narbonin, SI-CLP (stabilin-1 interacting chitinase-like protein), IDGF (imaginal disc growth factor), CFLE (cortical fragment-lytic enzyme) spore hydrolase, the type III and type V plant chitinases, the endo-beta-N-acetylglucosaminidases, and the chitolectins. The GH85 (glycosyl hydrolase, family 85) ENGases (endo-beta-N-acetylglucosaminidases) are closely related to the GH18 chitinases and are included in this alignment model. |
| 119360 | GH18_PF-ChiA-like | 2.40e-04 | 58 | 170 | 24 | 114 | PF-ChiA is an uncharacterized chitinase found in the hyperthermophilic archaeon Pyrococcus furiosus with a glycosyl hydrolase family 18 (GH18) catalytic domain as well as a cellulose-binding domain. Members of this domain family are found not only in archaea but also in eukaryotes and prokaryotes. PF-ChiA exhibits hydrolytic activity toward both colloidal and crystalline (beta/alpha) chitins at high temperature. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.09e-200 | 1 | 332 | 1 | 332 | |
| 4.45e-165 | 1 | 332 | 1 | 332 | |
| 8.19e-159 | 1 | 332 | 1 | 333 | |
| 3.88e-157 | 1 | 332 | 21 | 353 | |
| 9.98e-149 | 12 | 330 | 10 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.23e-59 | 33 | 331 | 8 | 291 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 2.29e-47 | 33 | 327 | 3 | 307 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 2.35e-47 | 33 | 327 | 4 | 308 | ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XTK_B ChiA1 from Aspergillus fumigatus in complex with acetazolamide [Aspergillus fumigatus A1163],2XUC_A Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_B Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XUC_C Natural product-guided discovery of a fungal chitinase inhibitor [Aspergillus fumigatus],2XVP_A ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163],2XVP_B ChiA1 from Aspergillus fumigatus, apostructure [Aspergillus fumigatus A1163] |
|
| 2.35e-47 | 33 | 327 | 4 | 308 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
|
| 1.15e-43 | 31 | 312 | 1 | 263 | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR [Hevea brasiliensis],1LLO_A Chain A, HEVAMINE [Hevea brasiliensis],2HVM_A Hevamine A At 1.8 Angstrom Resolution [Hevea brasiliensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.73e-132 | 29 | 335 | 33 | 348 | Endochitinase 2 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi2 PE=3 SV=2 |
|
| 9.54e-132 | 29 | 335 | 33 | 348 | Endochitinase 2 OS=Metarhizium anisopliae OX=5530 GN=chi2 PE=1 SV=1 |
|
| 4.60e-56 | 33 | 338 | 29 | 319 | Endochitinase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CTS1 PE=1 SV=2 |
|
| 2.84e-48 | 33 | 332 | 25 | 315 | Chitinase 3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CHT3 PE=1 SV=2 |
|
| 1.53e-46 | 2 | 308 | 4 | 281 | Acidic endochitinase SE2 OS=Beta vulgaris OX=161934 GN=SE2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
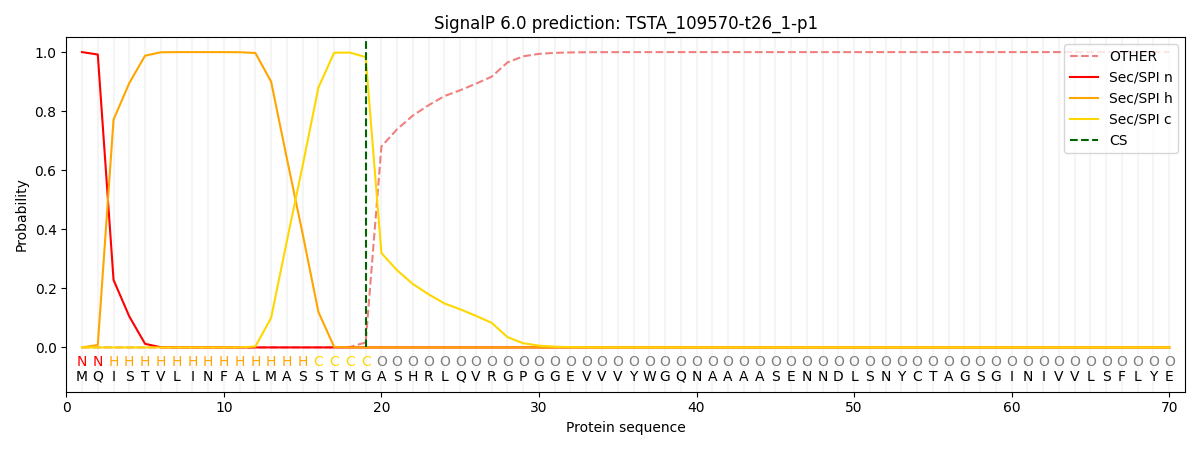
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000266 | 0.999699 | CS pos: 19-20. Pr: 0.9830 |
