You are browsing environment: FUNGIDB
CAZyme Information: TSTA_084670-t26_1-p1
You are here: Home > Sequence: TSTA_084670-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Talaromyces stipitatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces stipitatus | |||||||||||
| CAZyme ID | TSTA_084670-t26_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA5 | 1058 | 1559 | 3.8e-154 | 0.3747072599531616 |
| AA2 | 66 | 226 | 5.3e-22 | 0.6196078431372549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173829 | plant_peroxidase_like_1 | 2.46e-122 | 25 | 290 | 1 | 264 | Uncharacterized family of plant peroxidase-like proteins. This is a subgroup of heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX) which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. |
| 399910 | Glyoxal_oxid_N | 2.29e-25 | 1116 | 1312 | 59 | 243 | Glyoxal oxidase N-terminus. This family represents the N-terminus (approximately 300 residues) of a number of plant and fungal glyoxal oxidase enzymes. Glyoxal oxidase catalyzes the oxidation of aldehydes to carboxylic acids, coupled with reduction of dioxygen to hydrogen peroxide. It is an essential component of the extracellular lignin degradation pathways of the wood-rot fungus Phanerochaete chrysosporium. |
| 396406 | WSC | 2.42e-25 | 543 | 620 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 4.02e-25 | 646 | 723 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 173823 | plant_peroxidase_like | 1.73e-24 | 67 | 277 | 19 | 242 | Heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX), which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. Several sub-families can be identified. Class I includes intracellular peroxidases present in fungi, plants, archaea and bacteria, called catalase-peroxidases, that can exhibit both catalase and broad-spectrum peroxidase activities depending on the steady-state concentration of hydrogen peroxide. Catalase-peroxidases are typically comprised of two homologous domains that probably arose via a single gene duplication event. Class II includes ligninase and other extracellular fungal peroxidases, while class III is comprised of classic extracellular plant peroxidases, like horseradish peroxidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1565 | 1 | 1706 | |
| 0.0 | 537 | 1565 | 146 | 1118 | |
| 0.0 | 540 | 1565 | 81 | 1052 | |
| 0.0 | 540 | 1565 | 140 | 1109 | |
| 0.0 | 543 | 1565 | 154 | 1124 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.64e-17 | 1353 | 1544 | 413 | 603 | W288A mutant of GlxA from Streptomyces lividans: Cu-bound form [Streptomyces lividans TK24] |
|
| 8.75e-17 | 1353 | 1544 | 418 | 608 | W288A mutant of GlxA from Streptomyces lividans: apo form [Streptomyces lividans 1326],5LQI_B W288A mutant of GlxA from Streptomyces lividans: apo form [Streptomyces lividans 1326] |
|
| 8.89e-17 | 1353 | 1544 | 424 | 614 | Structure of Galactose Oxidase homologue from Streptomyces lividans [Streptomyces lividans],4UNM_B Structure of Galactose Oxidase homologue from Streptomyces lividans [Streptomyces lividans] |
|
| 1.08e-12 | 1121 | 1544 | 223 | 637 | Glactose oxidase C383S mutant identified by directed evolution [Fusarium sp.] |
|
| 1.08e-12 | 1121 | 1544 | 223 | 637 | Chain A, Galactose oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 643 | 1565 | 63 | 924 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 3.57e-262 | 1 | 730 | 5 | 715 | WSC domain-containing protein ARB_07870 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07870 PE=1 SV=1 |
|
| 6.93e-54 | 1096 | 1544 | 121 | 593 | Putative aldehyde oxidase Art an 7 OS=Artemisia annua OX=35608 PE=1 SV=1 |
|
| 4.17e-51 | 1061 | 1544 | 15 | 521 | Aldehyde oxidase GLOX OS=Vitis pseudoreticulata OX=231512 GN=GLOX PE=2 SV=1 |
|
| 1.38e-44 | 1116 | 1544 | 169 | 614 | Aldehyde oxidase GLOX1 OS=Arabidopsis thaliana OX=3702 GN=GLOX1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
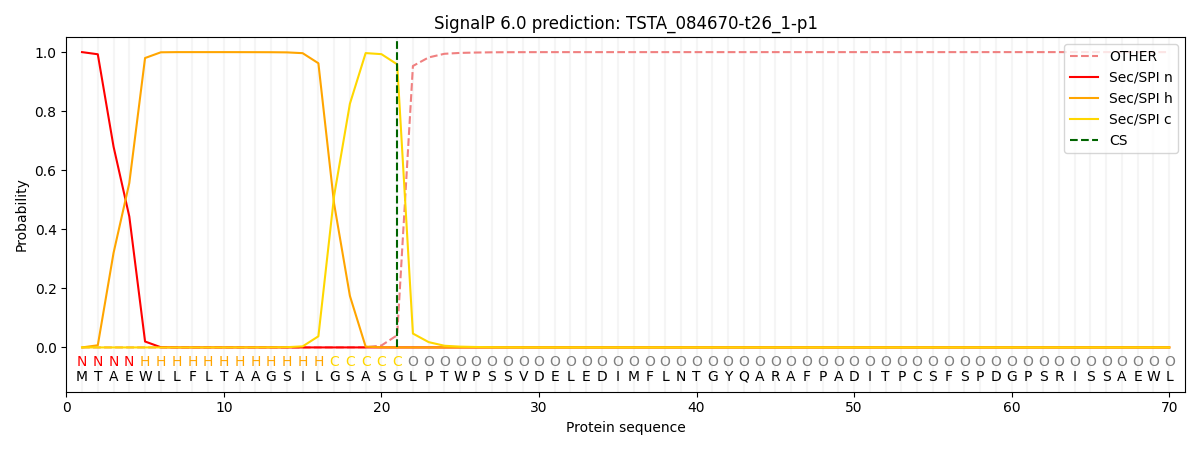
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000256 | 0.999685 | CS pos: 21-22. Pr: 0.9587 |
