You are browsing environment: FUNGIDB
CAZyme Information: TSTA_010130-t26_1-p1
You are here: Home > Sequence: TSTA_010130-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Talaromyces stipitatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces stipitatus | |||||||||||
| CAZyme ID | TSTA_010130-t26_1-p1 | |||||||||||
| CAZy Family | GT61 | |||||||||||
| CAZyme Description | endoglucanase-5 precursor, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 409015 | DPBB_GH45_endoglucanase | 5.27e-17 | 20 | 156 | 7 | 149 | double-psi beta-barrel fold of glycoside hydrolase family 45 endoglucanase EG27II and similar proteins. This group is made up of endoglucanases from mollusks similar to Ampullaria crossean endoglucanase EG27II, a glycoside hydrolase family 45 (GH45) subfamily B protein. Endoglucanases (EC 3.2.1.4) catalyze the endohydrolysis of (1-4)-beta-D-glucosidic linkages in cellulose, lichenin, and cereal beta-D-glucans. Animal cellulases, such as endoglucanase EG27II, have great potential for industrial applications such as bioethanol production. GH45 endoglucanases from mollusks adopt a double-psi beta-barrel (DPBB) fold. |
| 395595 | CBM_1 | 4.26e-12 | 292 | 320 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 3.19e-10 | 291 | 324 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.46e-149 | 1 | 327 | 1 | 338 | |
| 8.90e-107 | 1 | 324 | 1 | 251 | |
| 3.77e-98 | 1 | 328 | 1 | 282 | |
| 9.74e-98 | 1 | 325 | 1 | 248 | |
| 1.01e-94 | 1 | 324 | 1 | 246 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.43e-27 | 20 | 170 | 25 | 181 | Crystal structure of GH45 endoglucanase EG27II in apo-form [Ampullaria crossean],5XBX_A Crystal structure of GH45 endoglucanase EG27II in complex with cellobiose [Ampullaria crossean],5XC4_A Crystal structure of GH45 endoglucanase EG27II at pH4.0, in complex with cellobiose [Ampullaria crossean],5XC8_A Crystal structure of GH45 endoglucanase EG27II at pH5.5, in complex with cellobiose [Ampullaria crossean],5XC9_A Crystal structure of GH45 endoglucanase EG27II at pH8.0, in complex with cellobiose [Ampullaria crossean] |
|
| 3.50e-26 | 20 | 170 | 25 | 181 | Crystal structure of GH45 endoglucanase EG27II D137A mutant in complex with cellobiose [Ampullaria crossean] |
|
| 7.93e-21 | 20 | 166 | 17 | 167 | Chain A, ENDOGLUCANASE [Mytilus edulis] |
|
| 4.65e-10 | 290 | 324 | 2 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 8.74e-10 | 293 | 324 | 5 | 36 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.55e-92 | 1 | 324 | 1 | 241 | Endoglucanase-5 OS=Hypocrea jecorina OX=51453 GN=egl5 PE=3 SV=1 |
|
| 4.08e-20 | 20 | 166 | 17 | 167 | Endoglucanase OS=Mytilus edulis OX=6550 PE=1 SV=1 |
|
| 1.17e-10 | 279 | 324 | 460 | 505 | Exoglucanase 1 OS=Trichoderma harzianum OX=5544 GN=cbh1 PE=1 SV=1 |
|
| 1.20e-09 | 280 | 324 | 470 | 514 | Exoglucanase 1 OS=Hypocrea rufa OX=5547 GN=cbh1 PE=3 SV=2 |
|
| 2.01e-09 | 288 | 324 | 18 | 54 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
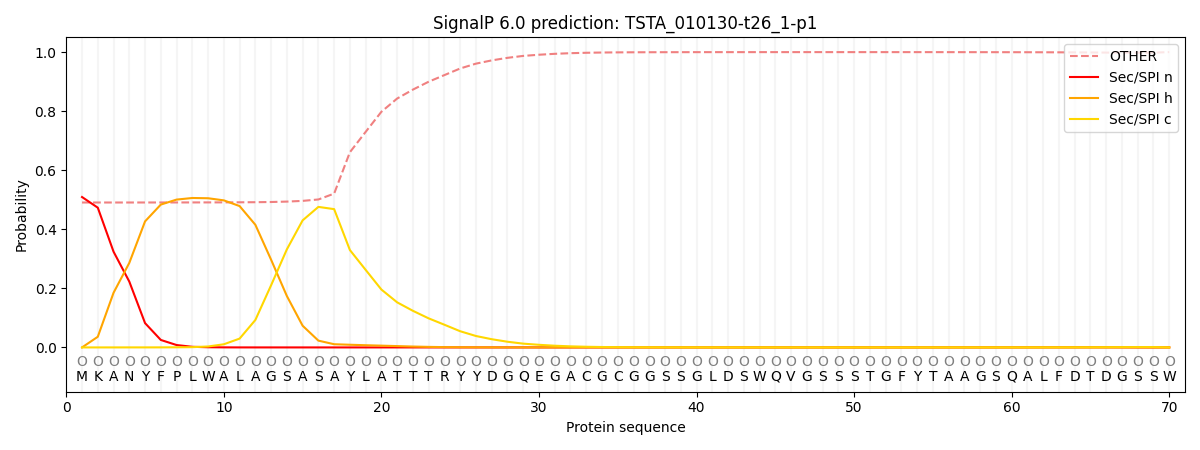
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.512759 | 0.487220 |
