You are browsing environment: FUNGIDB
CAZyme Information: TSTA_009910-t26_1-p1
You are here: Home > Sequence: TSTA_009910-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Talaromyces stipitatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces stipitatus | |||||||||||
| CAZyme ID | TSTA_009910-t26_1-p1 | |||||||||||
| CAZy Family | GT62 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 341045 | MFS_Azr1_MDR_like | 7.81e-24 | 461 | 593 | 111 | 315 | Saccharomyces cerevisiae Azole resistance protein 1 (Azr1p), and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This subfamily is composed of multidrug resistance (MDR) transporters including various Saccharomyces cerevisiae proteins such as azole resistance protein 1 (Azr1p), vacuolar basic amino acid transporter 1 (Vba1p), vacuolar basic amino acid transporter 5 (Vba5p), and Sge1p (also known as Nor1p, 10-N-nonyl acridine orange resistance protein, and crystal violet resistance protein). MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. This subfamily belongs to the Methylenomycin A resistance protein (also called MMR peptide) and similar multidrug resistance (MDR) transporters (MMR-like MDR transporter) family of the Major Facilitator Superfamily (MFS) of transporters. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 340868 | MFS_TRI12_like | 1.63e-13 | 484 | 677 | 301 | 506 | Fungal trichothecene efflux pump (TRI12) of the Major Facilitator Superfamily of transporters. This family includes Fusarium sporotrichioides trichothecene efflux pump (TRI12), which may play a role in F. sporotrichioides self-protection against trichothecenes. TRI12 belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 340880 | MFS_ARN_like | 8.04e-11 | 470 | 676 | 311 | 514 | Yeast ARN family of Siderophore iron transporters and similar proteins of the Major Facilitator Superfamily. The ARN family of siderophore iron transporters includes ARN1 (or ferrichrome permease), ARN2 (or triacetylfusarinine C transporter 1 or TAF1), ARN3 (or siderophore iron transporter 1 or SIT1 or ferrioxamine B permease) and ARN4 (or Enterobactin permease or ENB1). They specifically recognize siderophore-iron chelates are expressed under conditions of iron deprivation. They facilitate the uptake of both hydroxamate- and catecholate-type siderophores. This group also includes glutathione exchanger 1 (Gex1p) and Gex2p, which are proton/glutathione antiporters that import glutathione from the vacuole and exports it through the plasma membrane. The ARN family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 340879 | MFS_MMR_MDR_like | 2.37e-10 | 470 | 672 | 184 | 368 | Methylenomycin A resistance protein (also called MMR peptide) and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This family is composed of bacterial, fungal, and archaeal multidrug resistance (MDR) transporters including several proteins from Bacilli such as methylenomycin A resistance protein (also called MMR peptide), tetracycline resistance protein (TetB), and lincomycin resistance protein LmrB, as well as fungal proteins such as vacuolar basic amino acid transporters, which are involved in the transport into vacuoles of the basic amino acids histidine, lysine, and arginine in Saccharomyces cerevisiae, and aminotriazole/azole resistance proteins. MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. For example, MMR confers resistance to the epoxide antibiotic methylenomycin while TetB resistance to tetracycline by an active tetracycline efflux. MMR-like MDR transporters belong to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 369468 | MFS_1 | 3.63e-05 | 492 | 590 | 248 | 345 | Major Facilitator Superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.66e-167 | 1 | 430 | 1 | 445 | |
| 3.66e-167 | 1 | 430 | 1 | 445 | |
| 6.13e-164 | 18 | 430 | 20 | 449 | |
| 1.97e-142 | 94 | 428 | 1 | 326 | |
| 1.11e-141 | 94 | 428 | 1 | 326 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.15e-73 | 484 | 693 | 364 | 573 | MFS-type transporter ucsD OS=Acremonium sp. OX=2046025 GN=ucsD PE=3 SV=1 |
|
| 6.24e-49 | 480 | 692 | 327 | 540 | Efflux pump FUS6 OS=Gibberella fujikuroi (strain CBS 195.34 / IMI 58289 / NRRL A-6831) OX=1279085 GN=FUS6 PE=2 SV=1 |
|
| 7.14e-49 | 480 | 692 | 335 | 548 | Efflux pump FUS6 OS=Gibberella moniliformis (strain M3125 / FGSC 7600) OX=334819 GN=FUS6 PE=3 SV=1 |
|
| 9.48e-45 | 484 | 705 | 326 | 544 | MFS efflux transporter aclA OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=aclA PE=3 SV=1 |
|
| 3.99e-44 | 480 | 690 | 338 | 549 | MFS-type efflux pump LUC4 OS=Fusarium sp. OX=29916 GN=LUC4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
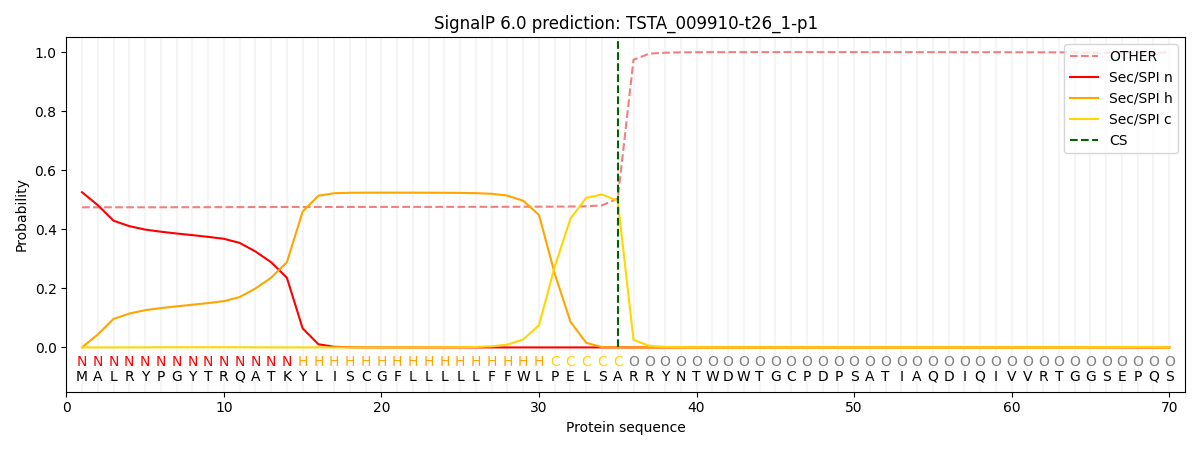
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.490934 | 0.509065 | CS pos: 35-36. Pr: 0.4962 |
TMHMM Annotations download full data without filtering help

| Start | End |
|---|---|
| 12 | 31 |
| 484 | 506 |
| 513 | 531 |
| 541 | 563 |
| 576 | 598 |
| 653 | 672 |
