You are browsing environment: FUNGIDB
CAZyme Information: TRIVIDRAFT_8282-t45_1-p1
You are here: Home > Sequence: TRIVIDRAFT_8282-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma virens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma virens | |||||||||||
| CAZyme ID | TRIVIDRAFT_8282-t45_1-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | glycoside hydrolase family 11 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 59 | 226 | 4.8e-57 | 0.96045197740113 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 3.05e-82 | 55 | 225 | 1 | 174 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.30e-165 | 1 | 231 | 1 | 231 | |
| 3.30e-165 | 1 | 231 | 1 | 231 | |
| 1.67e-150 | 1 | 231 | 1 | 231 | |
| 1.22e-131 | 1 | 231 | 1 | 226 | |
| 3.09e-107 | 1 | 221 | 1 | 224 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.87e-87 | 57 | 231 | 4 | 178 | Chain A, Endo-1,4-beta-xylanase I [Trichoderma reesei] |
|
| 3.21e-49 | 66 | 230 | 48 | 215 | Crystal structure of environmental isolated GH11 in complex with xylobiose and feruloyl-arabino-xylotriose [Escherichia coli] |
|
| 3.31e-49 | 66 | 230 | 48 | 215 | Environmentally isolated GH11 xylanase [Escherichia coli] |
|
| 6.06e-49 | 66 | 231 | 48 | 216 | Thermostable mutant of ENVIRONMENTALLY ISOLATED GH11 XYLANASE [Escherichia coli] |
|
| 8.97e-48 | 65 | 230 | 23 | 190 | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) [Talaromyces funiculosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.32e-94 | 1 | 231 | 1 | 229 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_74223 PE=1 SV=1 |
|
| 4.32e-94 | 1 | 231 | 1 | 229 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=xyn1 PE=1 SV=1 |
|
| 7.17e-73 | 57 | 221 | 44 | 210 | Endo-1,4-beta-xylanase B OS=Talaromyces funiculosus OX=28572 GN=xynB PE=1 SV=1 |
|
| 1.55e-57 | 1 | 229 | 1 | 230 | Probable endo-1,4-beta-xylanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnA PE=3 SV=1 |
|
| 2.94e-55 | 1 | 225 | 1 | 227 | Probable endo-1,4-beta-xylanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xlnA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
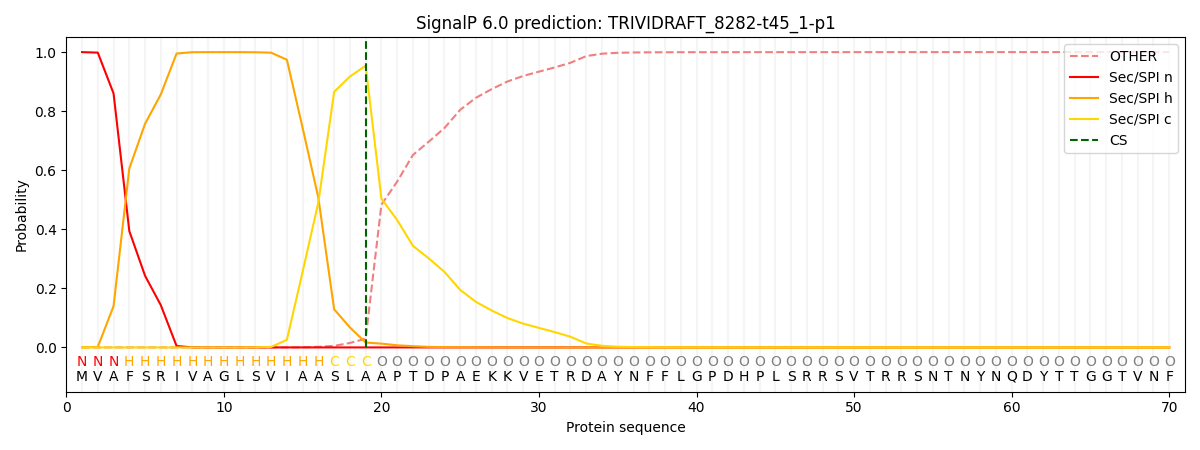
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000881 | 0.999103 | CS pos: 19-20. Pr: 0.9540 |
