You are browsing environment: FUNGIDB
CAZyme Information: TRIVIDRAFT_51554-t45_1-p1
Basic Information
help
| Species |
Trichoderma virens
|
| Lineage |
Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma virens
|
| CAZyme ID |
TRIVIDRAFT_51554-t45_1-p1
|
| CAZy Family |
GH3 |
| CAZyme Description |
glycoside hydrolase family 5 protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 478 |
ABDF02000075|CGC2 |
52165.76 |
6.6052 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_TvirensGv29-8 |
12405 |
413071 |
0 |
12405
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH30 |
25 |
470 |
1.7e-139 |
0.9956140350877193 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 405300
|
Glyco_hydr_30_2 |
1.29e-13 |
23 |
252 |
3 |
224 |
O-Glycosyl hydrolase family 30. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
478 |
1 |
478 |
| 0.0 |
1 |
478 |
1 |
478 |
| 0.0 |
1 |
478 |
1 |
478 |
| 2.52e-303 |
1 |
478 |
1 |
479 |
| 3.05e-290 |
1 |
478 |
1 |
478 |
TRIVIDRAFT_51554-t45_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.48e-304 |
1 |
478 |
1 |
479 |
Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
This protein is predicted as SP
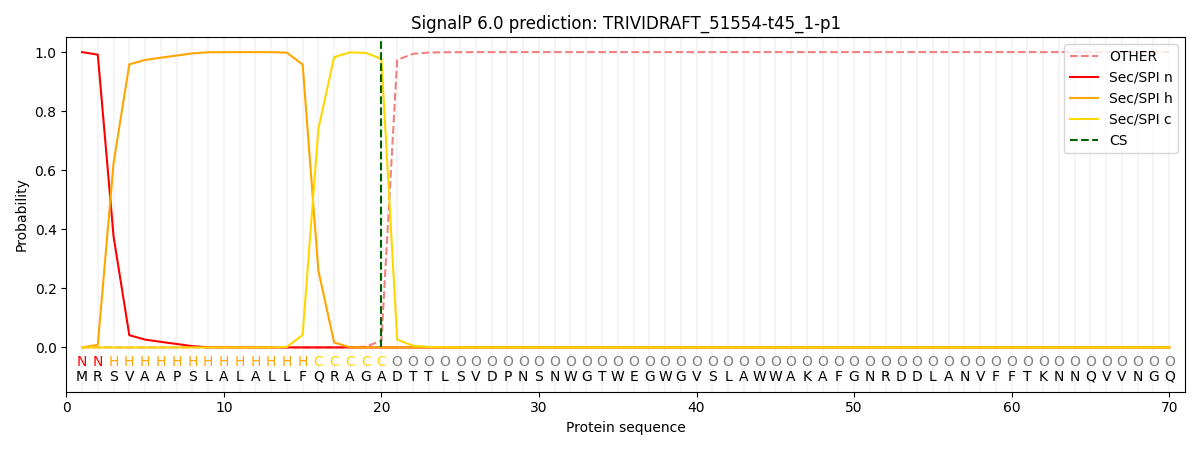
| Other |
SP_Sec_SPI |
CS Position |
| 0.000239 |
0.999725 |
CS pos: 20-21. Pr: 0.9775 |
There is no transmembrane helices in TRIVIDRAFT_51554-t45_1-p1.

