You are browsing environment: FUNGIDB
CAZyme Information: TRIVIDRAFT_32495-t45_1-p1
You are here: Home > Sequence: TRIVIDRAFT_32495-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma virens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma virens | |||||||||||
| CAZyme ID | TRIVIDRAFT_32495-t45_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | glycosyltransferase family 32 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT32 | 94 | 178 | 1e-19 | 0.9111111111111111 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226297 | OCH1 | 8.61e-24 | 53 | 299 | 54 | 310 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 398274 | Gly_transf_sug | 1.09e-11 | 89 | 179 | 2 | 89 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 239073 | Peptidase_C39_like | 0.009 | 237 | 288 | 12 | 64 | Peptidase family C39 mostly contains bacteriocin-processing endopeptidases from bacteria. The cysteine peptidases in family C39 cleave the "double-glycine" leader peptides from the precursors of various bacteriocins (mostly non-lantibiotic). The cleavage is mediated by the transporter as part of the secretion process. Bacteriocins are antibiotic proteins secreted by some species of bacteria that inhibit the growth of other bacterial species. The bacteriocin is synthesized as a precursor with an N-terminal leader peptide, and processing involves removal of the leader peptide by cleavage at a Gly-Gly bond, followed by translocation of the mature bacteriocin across the cytoplasmic membrane. Most endopeptidases of family C39 are N-terminal domains in larger proteins (ABC transporters) that serve both functions. The proposed protease active site is not conserved in all sub-families. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.97e-308 | 1 | 405 | 1 | 405 | |
| 2.97e-308 | 1 | 405 | 1 | 405 | |
| 5.82e-291 | 1 | 405 | 1 | 405 | |
| 2.84e-269 | 1 | 405 | 1 | 412 | |
| 8.14e-269 | 1 | 405 | 1 | 412 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.73e-39 | 72 | 337 | 142 | 393 | Initiation-specific alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=och1 PE=1 SV=2 |
|
| 1.25e-32 | 72 | 335 | 99 | 374 | Initiation-specific alpha-1,6-mannosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=OCH1 PE=3 SV=1 |
|
| 7.19e-31 | 72 | 317 | 130 | 370 | Putative glycosyltransferase HOC1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HOC1 PE=1 SV=3 |
|
| 1.60e-25 | 72 | 276 | 96 | 357 | Initiation-specific alpha-1,6-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=OCH1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
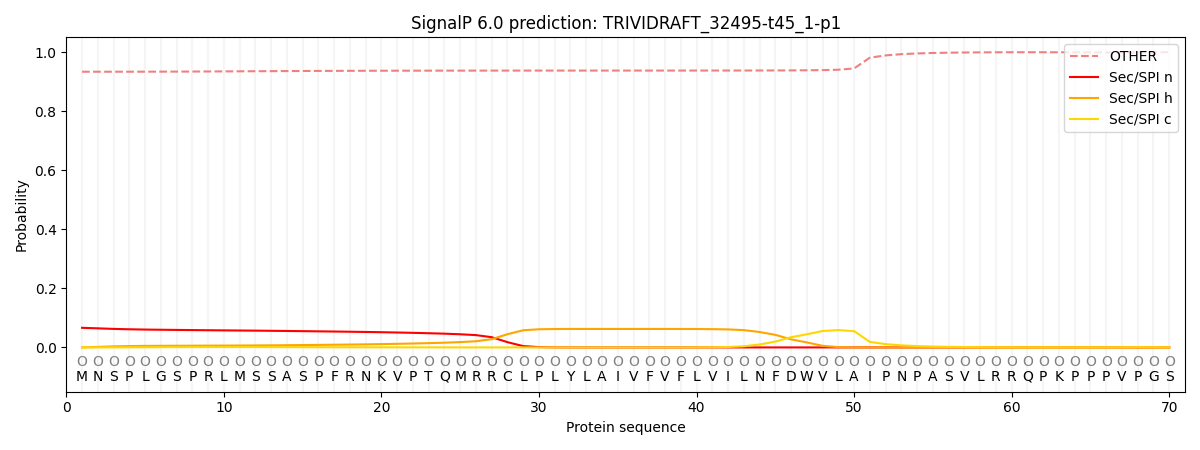
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.936948 | 0.063050 |

