You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_77547-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_77547-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
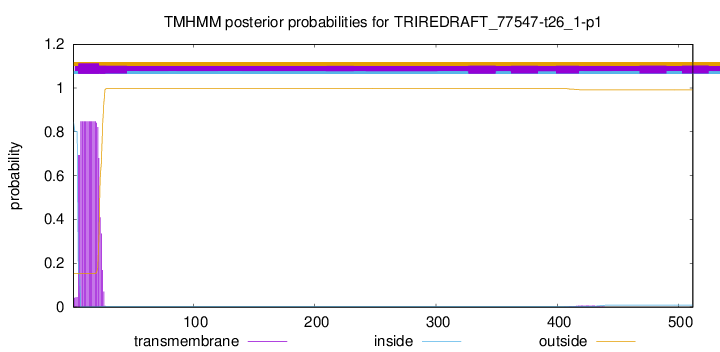
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_77547-t26_1-p1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | glycosyltransferase family 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 321 | 509 | 5.8e-19 | 0.4005235602094241 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 5.82e-23 | 156 | 507 | 94 | 404 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 224732 | YjiC | 5.54e-16 | 199 | 511 | 114 | 402 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 278624 | UDPGT | 1.09e-05 | 262 | 489 | 215 | 420 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 178170 | PLN02555 | 3.41e-04 | 345 | 496 | 296 | 454 | limonoid glucosyltransferase |
| 177813 | PLN02152 | 6.98e-04 | 403 | 496 | 343 | 438 | indole-3-acetate beta-glucosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.37e-295 | 1 | 512 | 1 | 512 | |
| 9.67e-288 | 1 | 512 | 1 | 512 | |
| 1.02e-246 | 1 | 511 | 1 | 458 | |
| 1.93e-231 | 1 | 512 | 2 | 514 | |
| 2.18e-159 | 1 | 508 | 1 | 510 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.31e-07 | 265 | 504 | 207 | 435 | Sterol 3-beta-glucosyltransferase (ugt51) from Saccharomyces cerevisiae (strain ATCC 204508 / S288c) [Saccharomyces cerevisiae S288C],5XVM_B Sterol 3-beta-glucosyltransferase (ugt51) from Saccharomyces cerevisiae (strain ATCC 204508 / S288c) [Saccharomyces cerevisiae S288C] |
|
| 2.38e-07 | 265 | 504 | 227 | 455 | Sterol 3-beta-glucosyltransferase (ugt51) from Saccharomyces cerevisiae (strain ATCC 204508 / S288c): UDPG complex [Saccharomyces cerevisiae S288C],5GL5_B Sterol 3-beta-glucosyltransferase (ugt51) from Saccharomyces cerevisiae (strain ATCC 204508 / S288c): UDPG complex [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.30e-76 | 38 | 504 | 7 | 502 | Glycosyltransferase sdnJ OS=Sordaria araneosa OX=573841 GN=sdnJ PE=1 SV=1 |
|
| 4.52e-60 | 34 | 507 | 3 | 487 | Glycosyltransferase buaB OS=Aspergillus burnettii OX=2508778 GN=buaB PE=3 SV=1 |
|
| 3.94e-13 | 376 | 504 | 433 | 558 | Erythritol-mannosyl-transferase 1 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=EMT1 PE=1 SV=1 |
|
| 3.73e-12 | 376 | 504 | 434 | 559 | Erythritol-mannosyl-transferase 1 OS=Pseudozyma antarctica (strain T-34) OX=1151754 GN=EMT1 PE=1 SV=1 |
|
| 3.85e-10 | 206 | 434 | 178 | 399 | UDP-glucuronosyltransferase 2B31 OS=Canis lupus familiaris OX=9615 GN=UGT2B31 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
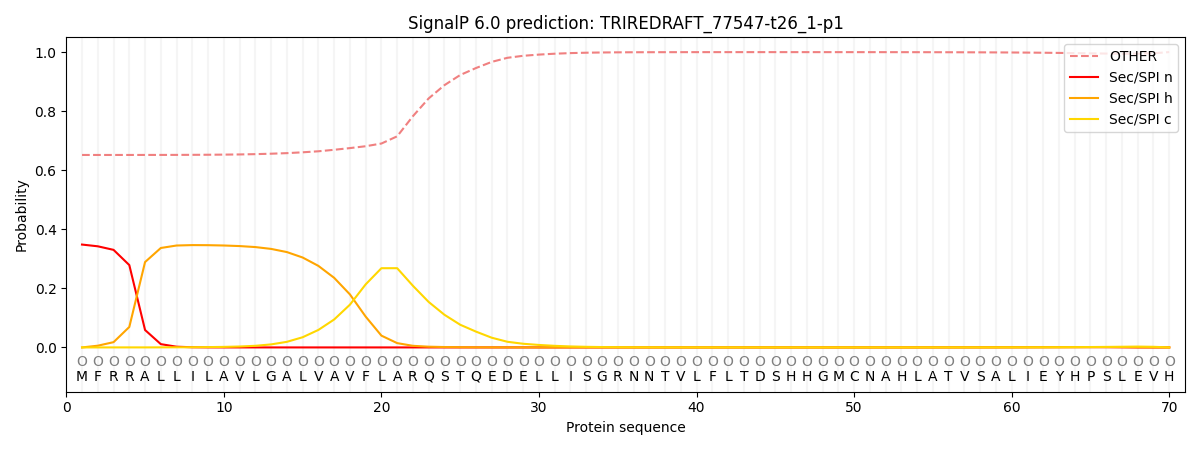
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.669131 | 0.330868 |