You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_74223-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_74223-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_74223-t26_1-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | glycoside hydrolase family 11 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 56 | 224 | 3.1e-61 | 0.96045197740113 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 6.94e-84 | 53 | 224 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.94e-165 | 1 | 229 | 1 | 229 | |
| 2.94e-165 | 1 | 229 | 1 | 229 | |
| 2.94e-165 | 1 | 229 | 1 | 229 | |
| 2.94e-165 | 1 | 229 | 1 | 229 | |
| 1.50e-158 | 1 | 229 | 1 | 229 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.49e-129 | 52 | 229 | 1 | 178 | Chain A, Endo-1,4-beta-xylanase I [Trichoderma reesei] |
|
| 1.83e-54 | 50 | 229 | 27 | 211 | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe [Aspergillus niger] |
|
| 2.26e-54 | 52 | 229 | 2 | 184 | Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger],1UKR_B Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger],1UKR_C Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger],1UKR_D Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger] |
|
| 3.20e-54 | 52 | 229 | 2 | 184 | Endo-1,4-Beta-Xylanase C [Aspergillus awamori],1T6G_C Chain C, Endo-1,4-beta-xylanase I [Aspergillus niger],1T6G_D Chain D, Endo-1,4-beta-xylanase I [Aspergillus niger] |
|
| 9.08e-54 | 52 | 229 | 2 | 184 | Chain A, Endo-1,4-beta-xylanase I [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.23e-166 | 1 | 229 | 1 | 229 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=TRIREDRAFT_74223 PE=1 SV=1 |
|
| 5.23e-166 | 1 | 229 | 1 | 229 | Endo-1,4-beta-xylanase 1 OS=Hypocrea jecorina (strain ATCC 56765 / BCRC 32924 / NRRL 11460 / Rut C-30) OX=1344414 GN=xyn1 PE=1 SV=1 |
|
| 1.34e-72 | 49 | 219 | 38 | 210 | Endo-1,4-beta-xylanase B OS=Talaromyces funiculosus OX=28572 GN=xynB PE=1 SV=1 |
|
| 1.82e-58 | 1 | 227 | 1 | 221 | Endo-1,4-beta-xylanase B OS=Phanerodontia chrysosporium OX=2822231 GN=xynB PE=1 SV=1 |
|
| 1.87e-57 | 1 | 227 | 1 | 227 | Endo-1,4-beta-xylanase xynf11a OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
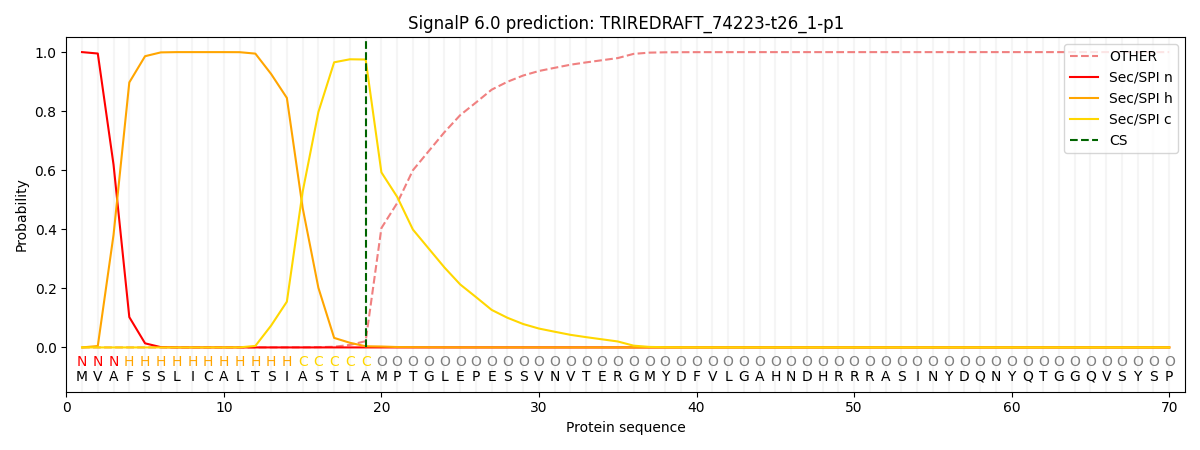
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000313 | 0.999638 | CS pos: 19-20. Pr: 0.9749 |
