You are browsing environment: FUNGIDB
CAZyme Information: TRIREDRAFT_73523-t26_1-p1
You are here: Home > Sequence: TRIREDRAFT_73523-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichoderma reesei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Hypocreaceae; Trichoderma; Trichoderma reesei | |||||||||||
| CAZyme ID | TRIREDRAFT_73523-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | carboxy terminal WSC domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA2 | 61 | 242 | 1.5e-23 | 0.6901960784313725 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 173829 | plant_peroxidase_like_1 | 2.60e-135 | 23 | 288 | 1 | 264 | Uncharacterized family of plant peroxidase-like proteins. This is a subgroup of heme-dependent peroxidases similar to plant peroxidases. Along with animal peroxidases, these enzymes belong to a group of peroxidases containing a heme prosthetic group (ferriprotoporphyrin IX) which catalyzes a multistep oxidative reaction involving hydrogen peroxide as the electron acceptor. The plant peroxidase-like superfamily is found in all three kingdoms of life and carries out a variety of biosynthetic and degradative functions. |
| 396406 | WSC | 2.47e-25 | 617 | 692 | 3 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 4.70e-25 | 831 | 908 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 4.99e-24 | 725 | 802 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 395089 | peroxidase | 8.62e-19 | 61 | 198 | 13 | 156 | Peroxidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-307 | 7 | 918 | 11 | 931 | |
| 1.14e-283 | 4 | 809 | 8 | 799 | |
| 1.12e-179 | 10 | 809 | 8 | 731 | |
| 3.17e-106 | 8 | 530 | 7 | 535 | |
| 1.33e-105 | 17 | 530 | 15 | 538 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.02e-09 | 615 | 703 | 91 | 182 | Wnt modulator Kremen in complex with DKK1 (CRD2) and LRP6 (PE3PE4) [Homo sapiens] |
|
| 3.49e-09 | 615 | 703 | 100 | 191 | Chain E, KRM1 [Homo sapiens],7BZU_E Chain E, KRM1 [Homo sapiens] |
|
| 3.54e-09 | 615 | 703 | 103 | 194 | Chain E, Kremen protein 1 [Homo sapiens] |
|
| 3.96e-09 | 615 | 703 | 131 | 222 | Wnt modulator Kremen crystal form I at 1.90A [Homo sapiens],5FWT_A Wnt modulator Kremen crystal form I at 2.10A [Homo sapiens],5FWU_A Wnt modulator Kremen crystal form II at 2.8A [Homo sapiens],5FWV_A Wnt modulator Kremen crystal form III at 3.2A [Homo sapiens] |
|
| 1.58e-07 | 68 | 258 | 29 | 236 | Crystal structure of chloroplastic ascorbate peroxidase from tobacco plants and structural insights for its instability [Nicotiana tabacum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.30e-175 | 15 | 809 | 15 | 715 | WSC domain-containing protein ARB_07870 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07870 PE=1 SV=1 |
|
| 1.59e-39 | 607 | 809 | 12 | 219 | Putative fungistatic metabolite OS=Chaetomium globosum (strain ATCC 6205 / CBS 148.51 / DSM 1962 / NBRC 6347 / NRRL 1970) OX=306901 GN=CHGG_05463 PE=1 SV=2 |
|
| 1.12e-30 | 606 | 922 | 57 | 408 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 5.49e-15 | 615 | 809 | 138 | 328 | WSC domain-containing protein 2 OS=Danio rerio OX=7955 GN=wscd2 PE=3 SV=1 |
|
| 2.03e-13 | 615 | 772 | 143 | 294 | WSC domain-containing protein 1 OS=Mus musculus OX=10090 GN=Wscd1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
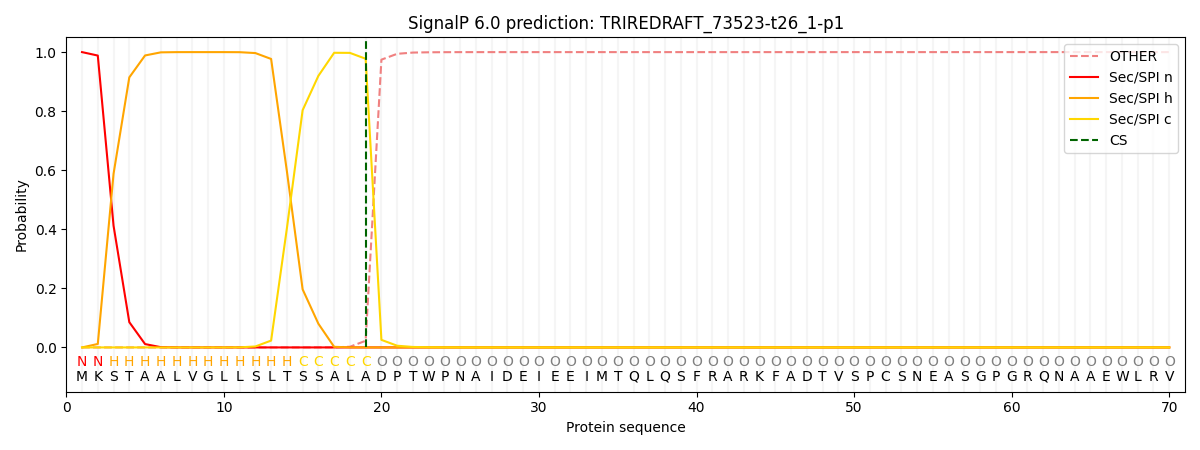
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000371 | 0.999600 | CS pos: 19-20. Pr: 0.9775 |
